Documentation Revision Date: 2024-10-02
Dataset Version: 1
Summary
This dataset includes 738 data files in cloud optimized GeoTIFF (*.tif) format.
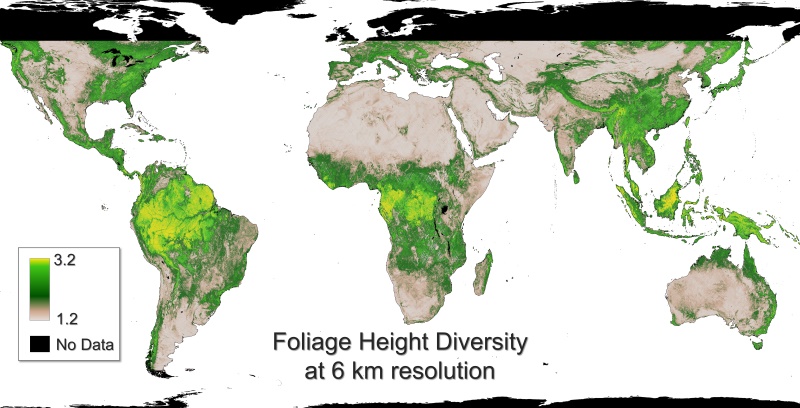
Figure 1. Mean foliage height diversity of GEDI shots acquired from April 2019 to March 2023 aggregated in 6-km grid cells.
Citation
Burns, P., C. Hakkenberg, and S.J. Goetz. 2024. Gridded GEDI Vegetation Structure Metrics and Biomass Density at Multiple Resolutions. ORNL DAAC, Oak Ridge, Tennessee, USA. https://doi.org/10.3334/ORNLDAAC/2339
Table of Contents
- Dataset Overview
- Data Characteristics
- Application and Derivation
- Quality Assessment
- Data Acquisition, Materials, and Methods
- Data Access
- References
Dataset Overview
This dataset consists of near-global, analysis-ready, multi-resolution vegetation structure metrics derived from NASA Global Ecosystem Dynamics Investigation (GEDI) Level 2 and 4A products associated with 25-m diameter lidar footprints. This dataset provides a comprehensive representation of near-global vegetation structure that is inclusive of the entire vertical profile, based solely on GEDI lidar, and validated with independent data. The GEDI sensor, mounted on the International Space Station (ISS), uses eight laser beams spaced by 60 m along-track and 600 m across-track on the Earth surface to measure ground elevation and vegetation structure between approximately 52 degrees North and South latitude. Between April 2019 to March 2023 (2019-04-17 to 2023-03-17), GEDI acquired 11 and 7.7 billion quality waveforms suitable for measuring ground elevation and vegetation structure, respectively. This dataset provides GEDI shot metrics aggregated into raster grids at three spatial resolutions: 1 km, 6 km, and 12 km. In addition to many of the standard L2 and L4A shot metrics, several additional metrics have been derived which may be particularly useful for applications in carbon and water cycling processes in earth system models, as well as forest management, biodiversity modeling, and habitat assessment. Variables include canopy height, canopy cover, plant area index, foliage height diversity, and plant area volume density at 5 m strata. Eight statistics are included for each GEDI shot metric: mean, bootstrapped standard error of the mean, median, standard deviation, interquartile range, 95th percentile, Shannon's diversity index, and shot count. Quality shot filtering methodology that aligns with the GEDI L4B Gridded Aboveground Biomass Density, Version 2.1 was used. In comparison to the current GEDI L3 dataset, this dataset provides additional gridded metrics at multiple spatial resolutions and over several temporal periods (annual and the full mission duration).
Related Publication:
Burns, P., C. Hakkenberg, and S. Goetz. 2024. Multi-resolution gridded maps of vegetation structure from GEDI. Submitted to Nature Scientific Data, 2024.
Related Data:
GEDI L2A
Dubayah, R., M. Hofton, J. Blair, J. Armston, H. Tang, and S. Luthcke. 2021. GEDI L2A Elevation and Height Metrics Data Global Footprint Level V002. NASA EOSDIS Land Processes Distributed Active Archive Center. https://doi.org/10.5067/GEDI/GEDI02_A.002
GEDI L2B
Dubayah, R., H. Tang, J. Armston, S. Luthcke, M. Hofton, and J. Blair. 2021. GEDI L2B Canopy Cover and Vertical Profile Metrics Data Global Footprint Level V002. NASA EOSDIS Land Processes Distributed Active Archive Center. https://doi.org/10.5067/GEDI/GEDI02_B.002
GEDI L3
Dubayah, R.O., S.B. Luthcke, T.J. Sabaka, J.B. Nicholas, S. Preaux, and M.A. Hofton. 2021. GEDI L3 Gridded Land Surface Metrics, Version 2. ORNL DAAC, Oak Ridge, Tennessee, USA. https://doi.org/10.3334/ORNLDAAC/1952
- Note: This is the standard GEDI mission product. This product has similar methodology but provides data at 1-km resolution and includes more GEDI metrics and aggregation statistics.
GEDI L4A
Dubayah, R.O., J. Armston, J.R. Kellner, L. Duncanson, S.P. Healey, P.L. Patterson, S. Hancock, H. Tang, J. Bruening, M.A. Hofton, J.B. Blair, and S.B. Luthcke. 2022. GEDI L4A Footprint Level Aboveground Biomass Density, Version 2.1. ORNL DAAC, Oak Ridge, Tennessee, USA. https://doi.org/10.3334/ORNLDAAC/2056
GEDI L4B
Dubayah, R.O., J. Armston, S.P. Healey, Z. Yang, P.L. Patterson, S. Saarela, G. Stahl, L. Duncanson, J.R. Kellner, J. Bruening, and A. Pascual. 2023. GEDI L4B Gridded Aboveground Biomass Density, Version 2.1. ORNL DAAC, Oak Ridge, Tennessee, USA. https://doi.org/10.3334/ORNLDAAC/2299
- Note: This is the standard GEDI mission product and provides gridded aboveground biomass density at 1-km resolution.
Acknowledgments:
Quality filtering criteria and the sub-orbit granule filter list (for product development) were made available by the GEDI Mission Science Team / UMD. These data are supported by NASA Terrestrial Ecology Grant Numbers NNL15AA03C and 80NSSC21K0189.
Data Characteristics
Spatial Coverage: Global within a nominal latitude extent of -52 to 52 degrees
Spatial Resolution: GEDI shot metrics are gridded to 1 km, 6 km, and 12 km
Temporal Coverage: 2019-04-17 to 2023-03-16
Temporal Resolution: Files cover six different time periods:
- Data acquired over the full mission, 2019-04-17 to 2023-03-16
- Data acquired in 2019, 2019-04-17 to 2019-12-31
- Data acquired in 2020, 2020-01-01 to 2020-12-31
- Data acquired in 2021, 2021-01-01 to 2021-12-31
- Data acquired in 2022, 2022-01-01 to 2022-12-31
- Data acquired in 2023, 2023-01-01 to 2023-03-16
Study Areas: Latitude and longitude are given in decimal degrees.
| Site | Westernmost Longitude | Easternmost Longitude | Northernmost Latitude | Southernmost Latitude |
|---|---|---|---|---|
| Global | -180.00 | 180.00 | 52.00 | -52.00 |
Data File Information
The dataset contains 738 raster files in Cloud-optimized GeoTIFF (*.tif) format. The files hold GEDI shot metrics (Table 1) at three spatial resolutions (1 km, 6 km, and 12 km) at six different time periods (see Temporal Resolution above). Most files contain eight bands reflecting statistics related to the shot metric (Table 2). Files with ‘counts’ metrics contain four bands (Table 3).
File Naming Convention
Files are named: gediv002_<metric>_<shot selection>_<start date>_<end date>_<spatial resolution>.tif where:
- <metric> is the GEDI derived metric characterizing elevation or vegetation characteristics. Many metrics include the suffix “-a0” indicating that the metric was derived using the default ground-finding algorithm. See Table 1 for metric descriptions.
- <shot selection>
- “ga” = all shots that are suitable for estimating elevation of the lowest mode (i.e. ground elevation). There are more “ga” shots relative to the other options since filtering requirements were less strict.
- “gf” = the first shots falling in a 30 m sub-grid that are suitable for estimating elevation of the lowest mode.
- “va” = all shots which are suitable for estimating vegetation metrics.
- “vf” = the (temporally) first shots falling in a 30-m sub-grid that are suitable for estimating vegetation metrics.
- <start date> and <end date> are the date (YYYYMMDD) range that shots were gridded over.
- <spatial resolution> is the pixel size in meters: 1000m, 6000m, or 12000m.
No data value: -9999
Number of bands: 8
Projection: WGS 84 / NSIDC EASE-Grid 2.0 Global (EPSG: 6933)
Resolution specific values
| Pixel Size (m) | Number of Rows | Number of Columns | Upper Left Corner X Map Coordinate | Upper Left Corner Y Map Coordinate |
|---|---|---|---|---|
| 1000 | 11553 | 34545 | -17272530.445 | 5776540.831 |
| 6000 | 1928 | 5759 | -17277530.445 | 5784540.831 |
| 12000 | 965 | 2881 | -17283530.445 | 5790540.831 |
Table 1. GEDI metrics, origin, and brief description.
| GEDI metric name | Original GEDI Product Level, Metric Name | Description |
|---|---|---|
| agbd-a0-qf | L4A, agbd | Predicted aboveground biomass density with the l4_quality_flag applied (Mg ha-1) |
| agbd-a0 | L4A, agbd | Predicted aboveground biomass density without the l4_quality_flag applied (Mg ha-1) |
| cover-a0 | L2B, cover | Total canopy cover, defined as the percent of the ground covered by the vertical projection of canopy material (unitless) |
| date-dec | L2A, derived from delta_time | Decimal date of acquisition (YYYY.nnnnn) |
| elev-lm-a0 | L2A, elev_lowestmode | Elevation of center of lowest mode (ground elevation) relative to WGS84 ellipsoid (meters) |
| even-pai-1m-a0 | Derived from L2B | Evenness of the L2B 1 m vertical Plant Area Index profile (m-1). Calculated as: fhd_normal / log(ceiling(rh100)) |
| even-pavd-5m-a0 | Derived from L2B PAVD profile | Evenness of the L2B 5 m vertical Plant Area Volume Density (PAVD) profile (m-1). Calculated as:If (rh-100-a0 > 5) { fhd-pavd-5m-a0 / log (number nonzero PAVD bins) } |
| fhd-pai-1m-a0 | L2B, fhd_normal | Foliage height diversity (FHD), or Shannon entropy index, calculated from 1-m vertical bins in the foliage profile, normalized by total plant area (PAI) index (unitless) |
| fhd-pavd-5m-a0 | Derived from L2B PAVD profile | FHD estimated from L2B 5 m plant area volume density (PAVD) vertical profile normalized by total PAVD (unitless) |
| num-modes-a0 | L2A, num_detectedmodes | Number of detected modes in rxwaveform (unitless) |
| pai-a0 | L2B, pai | Total Plant Area Index (PAI; m2 m-2) |
| pavd_0-5-frac | Derived from L2B PAVD profile | The fraction of PAVD in 0 to 5 m height bin relative to the sum of PAVD from all height bins (unitless) |
| pavd_0-5 | L2B, pavd_z | PAVD from 0 to 5 m (m2 m-3) |
| pavd_5-10 | L2B, pavd_z | PAVD from 5 to 10 m (m2 m-3) |
| pavd_10-15 | L2B, pavd_z | PAVD from 10 to 15 m (m2 m-3) |
| pavd_15-20 | L2B, pavd_z | PAVD from 15 to 20 m (m2 m-3) |
| pavd_20-25 | L2B, pavd_z | PAVD from 20 to 25 m (m2 m-3) |
| pavd_25-30 | L2B, pavd_z | PAVD from 25 to 30 m (m2 m-3) |
| pavd_30-35 | L2B, pavd_z | PAVD from 30 to 35 m (m2 m-3) |
| pavd_35-40 | L2B, pavd_z | PAVD from 35 to 40 m (m2 m-3) |
| pavd_40-45 | L2B, pavd_z | PAVD from 40 to 45 m (m2 m-3) |
| pavd_45-50 | L2B, pavd_z | PAVD from 45 to 50 m (m2 m-3) |
| pavd_50-55 | L2B, pavd_z | PAVD from 50 to 55 m (m2 m-3) |
| pavd_55-60 | L2B, pavd_z | PAVD from 55 to 60 m (m2 m-3) |
| pavd_60-65 | L2B, pavd_z | PAVD from 60 to 65 m (m2 m-3) |
| pavd_65-70 | L2B, pavd_z | PAVD from 65 to 70 m (m2 m-3) |
| pavd_70-75 | L2B, pavd_z | PAVD from 70 to 75 m (m2 m-3) |
| pavd_75-80 | L2B, pavd_z | PAVD from 75 to 80 m (m2 m-3) |
| pavd-bot-frac | Derived from L2B PAVD profile | Fraction of PAVD in the bottom half of the canopy relative to the sum of PAVD from all height bins (unitless). The midpoint is calculated as: (round((rh-100-a0/2)/5)*5)/5 |
| pavd-max-h | Derived from L2B PAVD profile | The upper height of the 5 m bin with maximum PAVD (m) |
| pavd-top-frac | Derived from L2B PAVD profile | Fraction of PAVD in the top half of the canopy relative to the sum of PAVD from all height bins (unitless). The midpoint is calculated as: (round((rh-100-a0/2)/5)*5)/5 |
| rh-50-a0 | L2A, rh | Relative height (RH) at the 50th percentile of returned energy; height of median energy (m) |
| rh-95-a0 | L2A, rh | RH at the 95th percentile of returned energy; a proxy for canopy height (m) |
| rh-98-a0 | L2A, rh | RH at the 98th percentile of returned energy; a proxy for canopy height (m) |
| rhvdr-b | Derived from L2A rh profile | Bottom canopy vertical distribution ratio (VDR; unitless). Calculated as:If (rh-100-a0 > 5 & rh-50-a0 >= 0 & rh-98-a0 >=0) { rh-50-a0 / rh-98-a0 } |
| rhvdr-m | Derived from L2A rh profile | Middle canopy VDR (unitless). Calculated as:If (rh-100-a0 > 5 & rh-25-a0 >= 0 & rh-75-a0 >= 0 & rh-98-a0 >=0) { (rh-75-a0 - rh-25-a0) / rh-98-a0 } |
| rhvdr-t | Derived from L2A rh profile | Top canopy VDR (unitless). Calculated as:If (rh-100-a0 > 5 & rh-50-a0 >=0 & rh-98-a0 >=0) { (rh-98-a0 - rh-50-a0) / rh-98-a0 } |
| sens-a0 | L2A, sensitivity | Maximum canopy cover that can be penetrated considering the SNR of the waveform (unitless) |
Table 2. Statistics used for per-pixel aggregation of GEDI shot metrics.
| Statistic Band Name Suffix | Description |
|---|---|
| mean | The mean of GEDI shot metric values within a pixel. |
| meanbse | Standard error of the mean calculated using bootstrap resampling. 100 bootstrap samples were created; each sample included 70% of shots, randomly selected. Standard error was calculated using the means of bootstrapped samples. (Only calculated when there are at least 10 GEDI shots in the grid cell.) |
| med | The median value (50th percentile) of GEDI shot metric values within a pixel. |
| sd | The standard deviation of GEDI shot metric values within a pixel. |
| iqr | The interquartile range (75 percentile minus 25th percentile) of GEDI shot metric values within a pixel. |
| p95 | The 95th percentile value of GEDI shot metric values within a pixel. |
| shan | Shannon’s diversity index (H) of GEDI shot metric values within a pixel. Calculated as:-1*(sum(p*log(p))) where p is the proportion of GEDI shot values per bin. For global map consistency, predefined GEDI metric bins were used (see Table 4). |
| countf | The count of GEDI shot metric values within a pixel. A 30-m sub-grid was used to select the (temporally) first GEDI shot acquired in each 30-m sub-grid cell. |
In addition, two shot count rasters were produced for each spatial resolution and temporal period. These rasters include the total number of shots that were suitable for gridding either all ground elevation (“ga”) or all vegetation metrics (“va”). In both cases, likely outliers were removed using the GEDI L4B excluded granules list (see companion file gedi_l4b_excluded_granules_v21.json in dataset Dubayah et al. 2023).
The four bands in the “counts” rasters are described in Table 3. They include per-pixel counts of unique shots, orbits, and tracks, as well as the average Nearest Neighbor Index (NNI; Evans et al. 2023), which is a proxy for quantifying spatial clustering/dispersion of GEDI shots. The NNI is expressed as the ratio of the observed Euclidean distance (m) divided by the expected distance for all shot pairs. The expected distance is the average distance between neighbors in a hypothetical random distribution. If the index is <1, the pattern exhibits spatial clustering; if the index is >1, shots are more evenly dispersed.
Table 3. “Counts” raster band descriptions.
| Statistic Band Name Suffix | Description |
|---|---|
| shots_count | The number of shots. For a shot to be counted in this layer the following fields need to be valid: longitude, latitude, elevation of the lowest mode, decimal date, and orbit. |
| orbits_uniq | The number of unique orbits. |
| tracks_uniq | The number of unique tracks. A track is the combination of orbit and beam. |
| shots_nni | The Nearest Neighbor Index (Evans et al. 2023). |
Companion File
This dataset contains one companion file: GEDI_Gridded_ALS_validation.pdf. This file contains methodological details associated with the validation procedure as well as validation plots and summary statistic tables for validation datasets.
Application and Derivation
The dataset provides gridded information on GEDI three-dimensional forest structure metrics at a near-global extent, which may be used to characterize carbon and water cycling processes in earth system models, as well as forest management, biodiversity modeling, and habitat assessment. GEDI uses a relatively sparse sampling design where laser shots from one orbital pass are spaced by approximately 60 m along-track and 600 m across-track on the Earth surface. At large extents, such as countries or continents, the billions of lidar shots (points/vectors) acquired by GEDI can be cumbersome to work with, and there are often large gaps between shots due to the ISS orbital geometry and cloud cover. Hence, several studies/datasets have applied different approaches to convert billions of GEDI shots to gridded maps (i.e. rasters). Potapov et al. (2021) used Landsat composites to predict GEDI canopy height (RH95) at 30-m spatial resolution over the GEDI domain, while Lang et al. (2023) used Sentinel-2 imagery to predict GEDI canopy height (RH98) at 10-m spatial resolution globally. These maps capture the spatial variation of forest canopy height at fine spatial resolution but lack information on the vertical profile, and have biases associated with optical satellite imagery saturation in dense canopies. The GEDI L3 Gridded Land Surface Metrics product (Dubayah et al. 2021) took a different approach. As the name implies, this product computes the mean and standard deviation (SD) of GEDI canopy height (RH100) and ground elevation at a spatial resolution of 1 km. In other words, if enough quality GEDI shots fall within a 1 km pixel, those shots are used to compute the mean and SD. Lastly, GEDI L4B Gridded Aboveground Biomass Density (Dubayah et al. 2023) used even higher quality GEDI shots in a hybrid inference framework to estimate the mean biomass (and standard error of the mean) at 1 km spatial resolution. Thus, there are currently several available gridded GEDI canopy height and biomass datasets. However, there are currently no gridded GEDI datasets associated with other GEDI L2 metrics, such as height of median energy, total plant area index, foliage height diversity, or vertical foliar profiles.
For this dataset, 26 metrics of interest from the GEDI L2 and L4A products and 10 custom metrics derived from the L2 dataset were gridded at three spatial resolutions: 1 km, 6 km, and 12 km. Multiple spatial resolutions were used to provide continuous (i.e. gap-free) coverage for different parts of the world. For example, with approximately four years of GEDI data, gridding at 1-km spatial resolution yields continuous spatial coverage for high latitudes (e.g. Washington State, USA) due to ISS orbital geometry, whereas continuous coverage in tropical forests is usually only possible at 12-km spatial resolution due to ISS orbital geometry and cloud cover. Furthermore, structural metrics were gridded for each year (2019, 2020, 2021, 2022, and 2023) of the GEDI mission as well as for the full mission (2019-04-17 to 2023-03-16) period. Gridding over the full mission period provides the most complete spatial coverage, but may not be appropriate for analyses in areas of coincident forest disturbance/loss.
A minimum of two shots per grid cell was required for gridding to occur, otherwise the pixel was assigned a nodata value (i.e. masked out). Two shots is the bare minimum for gridding using various statistics, but users should explore applying higher shot thresholds which may lead to more accurate gridded estimates (see Limitations and Recommendations section). The “countf” band in each gridded GEDI metric raster can be used to select pixels above a certain shot count threshold. Note that different filtering was used for gridding ground elevation (elev-lm-a0) versus vegetation structure metrics. The filtering details are provided below in Section 5, but the key difference is that additional filtering was applied for gridding vegetation structure metrics, requiring the highest geolocation accuracy, that the L2B algorithm was run, surface water percentage is less than 10%, the urban percentage is less than 50%, vegetation is “leaf-on”, no local outliers were detected, plant area index (PAI) and cover values fall within expected ranges, and the absolute elevation difference relative to the TanDEM-X DEM is 150 m or less.
Finally, in comparison with the GEDI L3 dataset, this approach goes beyond quantifying the mean and SD of GEDI shot metrics per pixel. There are four additional statistics for gridding: the median, interquartile range, 95th percentile, and Shannon’s diversity index (H). The median and interquartile range were computed since these metrics are less sensitive to outliers, compared to mean and SD. The 95th percentile statistic was computed to estimate the near-maximum metric value, ideally excluding the influence of outliers. While extensive filtering was used to select the highest quality GEDI shots, there is still a small percentage of outliers, usually associated with ground-finding errors in dense forest or low clouds/fog. Shannon’s H was also calculated as a measure of metric entropy. The SD, IQR, and Shannon’s H may also characterize horizontal forest structure heterogeneity of the pixel of interest (e.g. Carrasco et al. 2019; Torresani et al. 2023), especially if the pixel contains a large number of shots which are relatively well-distributed. In this regard, the number of GEDI shots per pixel and an estimate of shot spatial clustering/dispersion (“shots_nni” band in the count rasters) is provided.
Shannon Diversity Calculation
The Shannon Diversity index (H) was computed using a per-pixel histogram of GEDI shot metric values. However, given that Shannon’s Diversity index is sensitive to the total number of bins, for global consistency each metric's bin width was defined so that the total number of bins used for each metric was relatively equivalent. Specifically, for each metric, the range of the bulk (~95%) of the global distribution was identified and divided that number by 20 to determine the bin width. For example, the 95th percentile of the 1-km median pai_a0 raster is 5.6. Dividing 5.6 by 20 equals 0.28, which was rounded to 0.25. For some metrics, a slightly different bin size was chosen, informed by an understanding of the metric precision and/or the ecological relevance of a particular value (Table 4). For rh_98_a0 for example, the estimated bin size was 1.8 m, but this bin size was increased to 3 m given the GEDI’s long laser pulse width and potential of some canopies to reach >60 m (which would be ~20 bins). This empirical bin width determination ensures relative cross-compatibility among Shannon diversity values across metrics, while allowing for more than 20 bins (and hence higher Shannon's values) for those pixels exceeding the maximum of the 95th percentile of the global distribution.
Table 4. Histogram bin definitions for computing Shannon’s Diversity index across structural metrics.
| GEDI metric name | Histogram Min. | Histogram Max. | Bin Size |
|---|---|---|---|
| agbd-a0-qf | 0 | 8000 | 0.05 |
| agbd-a0 | 0 | 1 | 0.0833 |
| cover-a0 | 0 | 2023.5 | 200 |
| date-dec | 2019 | 9000 | 0.01 |
| elev-lm-a0 | -200 | 2 | 0.05 |
| even-pai-1m-a0 | 0 | 1 | /0.1 |
| even-pavd-5m-a0 | 0 | 4 | 0.1 |
| fhd-pai-1m-a0 | 0 | 4 | 1 |
| fhd-pavd-5m-a0 | 0 | 20 | 0.25 |
| num-modes-a0 | 1 | 12 | 0.025 |
| pai-a0 | 0 | 1 | 0.01 |
| pavd_0-5-frac | 0 | 1 | 0.01 |
| pavd_0-5 | 0 | 1 | 0.01 |
| pavd_5-10 | 0 | 1 | 0.01 |
| pavd_10-15 | 0 | 1 | 0.01 |
| pavd_15-20 | 0 | 1 | 0.01 |
| pavd_20-25 | 0 | 1 | 0.01 |
| pavd_25-30 | 0 | 1 | 0.01 |
| pavd_30-35 | 0 | 1 | 0.01 |
| pavd_35-40 | 0 | 1 | 0.01 |
| pavd_40-45 | 0 | 1 | 0.01 |
| pavd_45-50 | 0 | 1 | 0.01 |
| pavd_50-55 | 0 | 1 | 0.01 |
| pavd_55-60 | 0 | 1 | 0.01 |
| pavd_60-65 | 0 | 1 | 0.01 |
| pavd_65-70 | 0 | 1 | 0.01 |
| pavd_70-75 | 0 | 1 | 0.025 |
| pavd_75-80 | 0 | 1 | 5 |
| pavd-bot-frac | 0 | 100 | 0.025 |
| pavd-max-h | 0 | 1 | 1.5 |
| pavd-top-fr | 0 | 120 | 3 |
| ac | -99 | 120 | 3 |
| rh-50-a0 | -99 | 120 | 0.025 |
| rh-95-a0 | -99 | 1 | 0.025 |
| rh-98-a0 | 0 | 1 | 0.025 |
| rhvdr-b | 0 | 1 | 0 |
| rhvdr-m | 0 | 20 | 0 |
| rhvdr-t | 8000 | 20 | 0 |
Quality Assessment
Per-pixel accuracy was assessed by validating gridded pixels with independent airborne lidar scanning (ALS) surveys and uncertainty was estimated through bootstrapping of GEDI shots.
ALS validation
High-resolution gridded ALS data was used to validate select 1-km and 6-km gridded GEDI metrics. The following ALS datasets were used for validation: (1) National Ecological Observation Network (NEON, 2021) , USA (2) Sonoma County, CA, USA (3) Coconino National Forest, AZ, USA (4) NASA CMS Indonesia (Melendy et al., 2017), (5) EFForTS Indonesia (Camarretta and Schlund, 2021; Schlund et al., 2023), and (6) SAFE Malaysia (Swinfield et al., 2020). See the companion file GEDI_Gridded_ALS_validation.pdf for additional details.
User note: See the companion file GEDI_Gridded_ALS_validation.pdf for methodological details associated with the validation procedure as well as additional validation plots and summary statistic tables for validation datasets.
Per-pixel uncertainty from bootstrapping
Uncertainty of the per-pixel mean of each GEDI metric was estimated using a bootstrapping approach. For every pixel with at least 10 shots (at each resolution), 100 unique random samples of the shots falling within that pixel were taken. For each sample 70% of the available shots (without replacement) were randomly selected. The mean of each unique sample was calculated, followed by the bootstrap standard error of the estimated mean as recommended by Efron and Tibshirani (1994) and McRoberts et al. (2023) using the following bootstrap standard error equation:
meanbse = 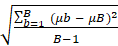 , where
, where
- b corresponds to an individual bootstrap
- B is the total number of bootstraps (100 in this case)
- μb is the per-pixel mean value of a GEDI metric associated with an individual bootstrap
- μB is the per-pixel mean of all individual bootstrap means, that is
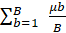
This method is designed to characterize the uncertainty associated with the GEDI sampling strategy, but it also incorporates uncertainty associated with variability of terrain and vegetation structure within each pixel.
Data Acquisition, Materials, and Methods
GEDI data download
GEDI L2A and L2B (version 2), as well as GEDI L4A (version 2.1) orbit granule files (.h5) from April 17 2019 to March 16 2023 were downloaded to Northern Arizona University’s (NAU) high performance computing system (HPC) (Table 5). L2 products were downloaded using a file list obtained from NASA Earthdata search. A shell script with the wget utility was used to automatically download each orbit granule file and verify that the checksum of the downloaded orbit granule file matched the checksum in the associated orbital granule XML file. Checksum verification was an important step considering intermittent connections to the LPDAAC Data Pool. The downloaded L2 data had an approximate volume of 126 Tb. Globus file transfer utility was used to automatically sync the L4A dataset from the ORNL DAAC. The downloaded L4A data had an approximate volume of 14 Tb.
Table 5. Summary of downloaded GEDI datasets
| GEDI dataset | Number of downloaded and checksum-matched orbit granules | Number of orbit granules available* | Size on Disk |
|---|---|---|---|
| L2A v2 | 74390 | 74811 | 103 Tb |
| L2B v2 | 74904 | 74810 | 23 Tb |
| L4A v2.1 | 75038 | 74860 | 14 Tb |
* Number of orbit granules available from NASA Earthdata Search as of Feb. 6 2024
Orbital processing and quality filtering
Most processing was done using a combination of R (R Core Team, 2021) and bash scripts. The SLURM Workload Manager on NAU’s HPC was used to distribute jobs. First, L2A, L2B, and L4A orbit granules were matched using a unique portion of their file names. Then, data were extracted from each product granule and converted to a R data table. The only metrics extracted were those associated with the default ground-finding algorithm (“a0”). A quality filtering recipe developed in collaboration with GEDI Science Team members from University of Maryland and NASA Goddard was used to identify the highest quality GEDI vegetation shots; this recipe follows the approach used for the GEDI L4B product.
Filtering
Below is pseudo-code for initial quality-filtering of each GEDI product. Initial filtering was used to select quality shots that are suitable for ground elevation and vegetation structure metrics.
L2A
L2A_filt = L2A[sens_a0 > 0.9 & sens_a0 <= 1
& sens_a2 > 0.9 & sens_a2 <= 1
& surf_flag == 1
& stale_flag == 0
& get(rh100) >= 0 & get(rh100) < 120
& get(ground_elev) > -200
& get(ground_elev) < 9000
& elev_diff_dem < 150 & elev_diff_dem > -150
& l2a_qflag_a0 == 1
& if(pft == 2 & region %in% c(4,5,6)){sens_a2 > 0.98} else {sens_a2 > 0.95}]
L2B
L2B_filt = L2B[sens_a0 > 0.9 & sens_a0 <= 1
& surf_flag == 1
& stale_flag == 0
& rh_100_a0 >= 0 & rh_100_a0 < 120
& elev_lm_a0 > -200
& elev_lm_a0 < 9000
& elev_diff_dem < 150 & elev_diff_dem > -150
& l2a_qflag_a0 == 1]
L4A
L4A_filt = L4A[sens_a0 > 0.9 & sens_a0 <= 1
& sens_a2 > 0.9 & sens_a2 <= 1
& surf_flag == 1
& stale_flag == 0
& elev_lm_a0 > -200
& elev_lm_a0 < 9000
& l2a_qflag_a0 == 1
& if(pft == 2 & region %in% c(4,5,6)){sens_a2 > 0.98} else {sens_a2 > 0.95}]
Initial filtered L2A, L2B, and L4A tables were joined together, matching by shot number, longitude of the lowest mode, and latitude of the lowest mode. Then, a dictionary of local outlier granules produced by University of Maryland was used to attribute orbit segments (“loc_out_umd”) as having local outliers (1) or not (0). That table is part of the GEDI L4B dataset (gedi_l4b_excluded_granules_v21.json; Dubayah et al. 2023).
Next, a L2 high-quality flag (“l2_hqflag”) was created to distinguish quality shots that are suitable for ground elevation (l2_hqflag == 0 | 1) versus those that are suitable for vegetation metrics (l2_hqflag == 1). To summarize, the L2 high-quality flag signifies the highest geolocation accuracy, that the L2B algorithm was run, surface water percentage is less than 10%, the urban percentage is less than 50%, vegetation is “leaf-on”, no local outliers were detected, PAI and cover values fall within expected ranges, and the absolute elevation difference relative to the TanDEM-X DEM is 150 m or less.
l2_hqflag = ifelse(deg_flag %in% c(0,3,8,10,13,18,20,23,28,30,33,38,40,43,48,60,63,68)
& l2b_algrun_flag == 1
& l2b_qflag_a0 == 1
& ls_waterp < 10
& urb_prop < 50
& !(leafoff_flag == 1)
& loc_out_umd == 0
& pai_a0 >= 0
& pai_l1 >= 0
& pavd_0_5 >= 0
& cover_a0 >= 0
& cover_a0 <= 1
& cover_l1 >= 0
& cover_l1 <= 1
& abs(elev_diff_dem) < 150,
1, 0)]
A L4A high-quality flag (“l4a_hqflag”) was created to distinguish high-quality shots that are suitable for aboveground biomass density (l4a_hqflag == 1) versus those that are suitable only for vegetation metrics (l4a_hqflag == 0 | 1). This flag was only used when gridding the metric agbd-a0-qf.
l4a_hqflag = ifelse(l2_hqflag == 1
& l4a_algrun_flag == 1
& l4a_qflag_a0 == 1
& agbd_a0 >= 0,
1, 0)]
Both the l2_hqflag and l4a_hqflag are used for filtering during the gridding procedure. Subsequently, the initial quality-filtered and joined shot tables (attributed with the l2_hqflag and l4a_hqflag flags) associated with each orbit granule were cropped to a regular 1x1 degree grid (EPSG 4326 geographic coordinates). In this process, the quality shots of each orbit granule were divided into 1x1 degree chunks, resulting in a large number of spatially-indexed tables
Gridding procedure
A separate gridding (SLURM) job was run for each 1x1 degree chunk. Initially, all quality shot chunk tables inside of and within a distance of 0.25 degrees of the 1x1 degree chunk of interest were combined. The inclusion of a 0.25-degree buffer was crucial to ensure that the edges of the gridded rasters matched, thereby eliminating edge artifacts in the final mosaic. This combination resulted in a data table, which was then used to select a specific GEDI metric of interest and filter according to a designated time period (annual or full mission). It was ensured that each quality-filtered shot contained valid values for key fields such as lon_lm_a0, lat_lm_a0, lon_lm_a0_6933, lat_lm_a0_6933, elev_lm_a0, date_dec, and orbit. A distinction was made between shots suitable for estimating ground elevation and those for assessing vegetation structure - the field l2_hqflag was employed to identify the highest quality shots for gridding vegetation structure metrics. Subsequently, a 30-m raster was created to select only the first shot within each 30-m grid cell. This step effectively reduced dense point clusters, aiding in the minimization of spatial biases in the gridded maps. The process concluded with a loop over each spatial resolution and time period, where each GEDI metric was gridded using the R function terra::rasterize, with specified functions for each aggregation statistic. A minimum of two shots per grid cell was required for gridding; otherwise, the pixel was assigned a nodata value and masked out.
For each temporal period, every 1x1 degree SLURM job generated 123 raster tiles. These tiles were then mosaicked together using GDAL (GDAL/OGR Contributors 2024), producing multi-resolution gridded rasters that span the entirety of the GEDI domain.
Limitations and Recommendations
Considering that this is a product with near-global extent, the uncertainty and validation assessments should be viewed as a work in progress. Validation assessments (see companion file GEDI_Gridded_ALS_validation.pdf) indicate that the gridding procedure performs well in areas with high GEDI shot density and low to moderate topographic relief, such as the NEON sites in the USA. The mean, median, and 95th percentile aggregation statistics show the best fit relative to corresponding gridded ALS. The SD, IQR, and Shannon’s H statistics generally have poorer fits and higher relative errors. These results suggest that GEDI generally does a good job of capturing the central and maximum tendencies of various forest structure metrics at multiple spatial resolutions, but does not always capture (horizontal) variability quite as well. The latter result is not necessarily surprising considering GEDI’s sampling density.
Users should be cautious when using this product in areas with low shot densities, like the tropics. ALS validation in Indonesia and Malaysia showed mixed results when using a minimum of two GEDI shots per grid cell. RH98 validated well in Kalimantan, Indonesia, but relatively poorly in Jambi, Indonesia and Sabah, Malaysia. The poor validation is likely the result of a combination of factors, including low shot density, erroneous GEDI measurements associated with clouds, and/or the forest dynamics in the region. Work is ongoing to estimate shot density thresholds that result in relatively accurate gridded estimates, but the impact of the minimum number of shots per grid cell on gridded GEDI metric accuracy is highlighted in Figure 2. RMSE decreases and model fit improves as the minimum number of shots per grid is increased. Although, the tradeoff of increasing the minimum number of shots per grid is that fewer grid cells will be available for analysis. Users may also find it beneficial to explore per-pixel filters using the associated “counts_vf” rasters which include the number of unique tracks and orbits per pixel. Requiring more than 1 orbit per pixel decreases the likelihood of errors associated with ground-finding and/or inclusion of low clouds in the returned waveform.
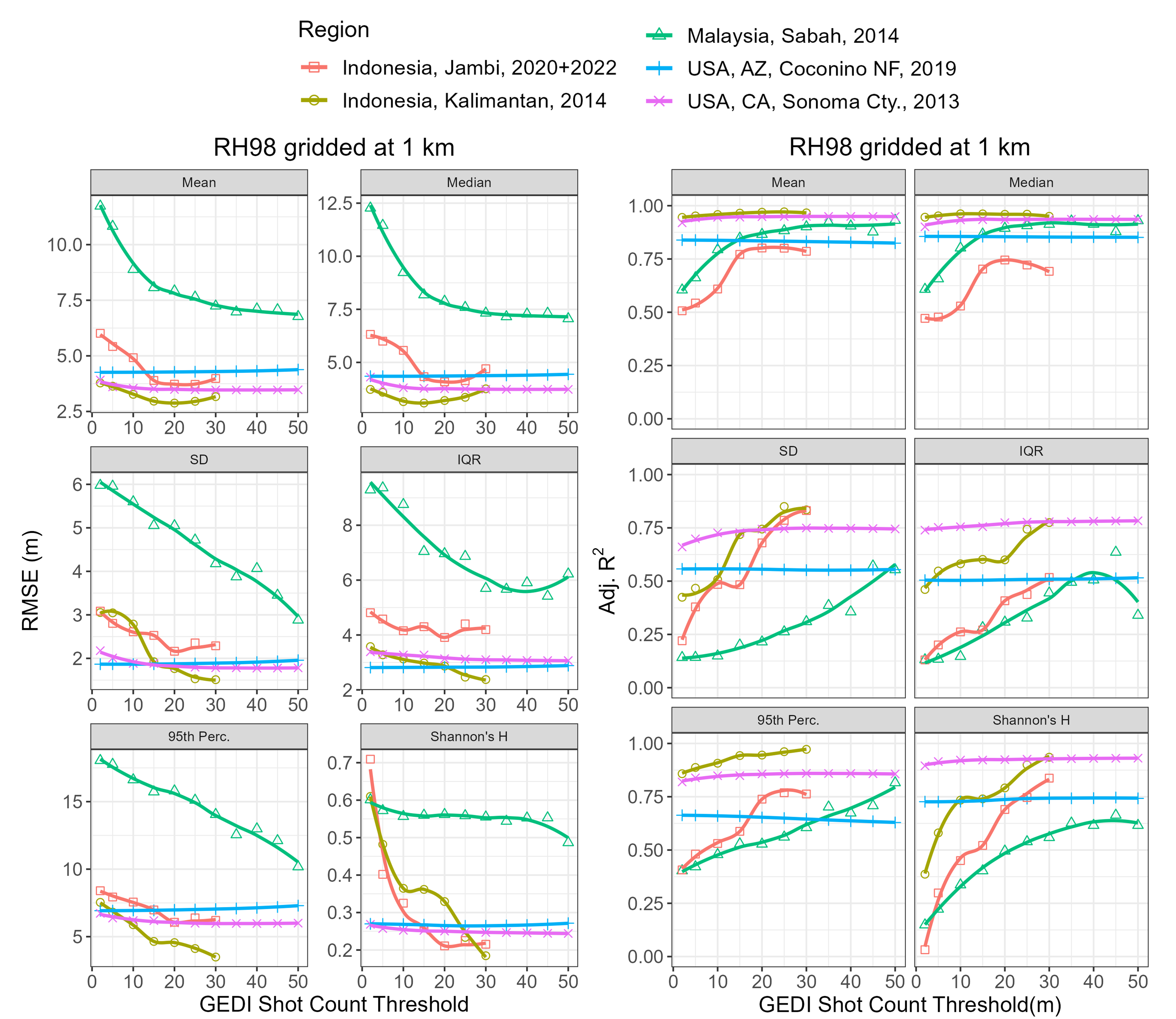
Figure 2. Comparison of airborne lidar survey (ALS) RH98 vs GEDI RH98 RMSE and adjusted R2 as a function of minimum number of GEDI shots per grid. The black line has a 1:1 relationship while the purple line corresponds to a linear fit (ALS ~ GEDI) of 1 km cells from all NEON sites.
Users should also be aware that topography influences many GEDI metrics and may produce artifacts in some gridded GEDI metrics. Since the GEDI laser pulse width is relatively long, the RH profile associated with the returned waveform may be further elongated on steep slopes or rough terrain, even if the area has little to no vegetation. An example of this topographic effect can be seen in the Grand Canyon of Arizona, USA (Figure 3) where vegetation within the canyon is generally low stature, yet 1 km gridded mean RH98 (an estimate of canopy height) frequently exceeds 10 m in this area.
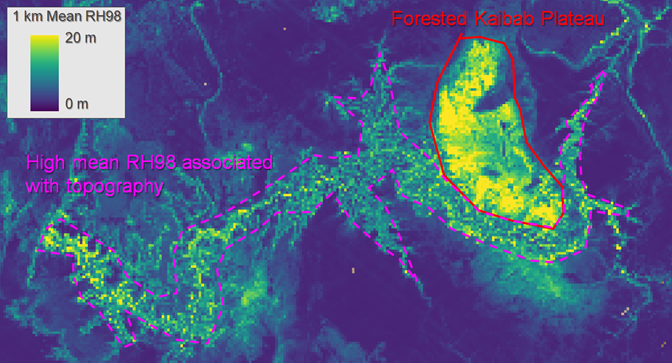
Figure 3 The influence of topography on 1-km mean GEDI RH98. High mean RH98 values correspond with Ponderosa Pine forest on the Kaibab Plateau, but also with steep/rough slopes of the Grand Canyon in Arizona, USA.
Finally, users may notice unexpected gaps in the mid- to high-latitudes, especially in 1-km resolution gridded rasters. Globally, most gaps are associated with ISS orbital geometry and cloud cover patterns, but there are some surprising gaps associated with vegetation phenology. Given that our primary goal was to produce gridded maps of vegetation structure metrics, “leaf-on” GEDI shots were used. The exact timing of “leaf-on” vs “leaf-off” was estimated using a VIIRS/NPP data product (VNP22Q2) which has its own uncertainties and limitations. Hence, some large regions containing deciduous vegetation have less gridded coverage relative to pixels at similar latitudes. Examples where gridded coverage is limited due to phenology include the Eastern USA and Sierra Madre Occidental, Mexico (Figure 4).
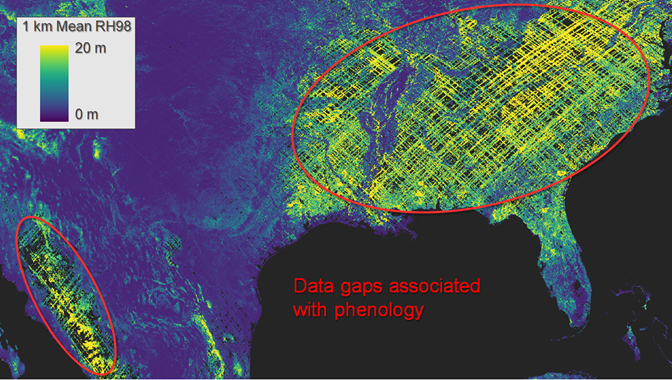
Figure 4. Examples of data gaps in mean GEDI RH98 from April 2019 to March 2023 gridded at 1 km. The gaps are the result of stringent quality filtering, phenology (leaf-off GEDI shots are excluded), coupled with ISS orbital geometry and cloud cover patterns.
Data Access
These data are available through the Oak Ridge National Laboratory (ORNL) Distributed Active Archive Center (DAAC).
Gridded GEDI Vegetation Structure Metrics and Biomass Density at Multiple Resolutions
Contact for Data Center Access Information:
- E-mail: uso@daac.ornl.gov
- Telephone: +1 (865) 241-3952
References
Burns, P., C. Hakkenberg, and S. Goetz. 2024. Multi-resolution gridded maps of vegetation structure from GEDI. Submitted to Nature Scientific Data, 2024.
Camarretta, N. and M. Schlund. 2021. Canopy Height Models. GRO.data, V2. https://doi.org/10.25625/CKLY7X
Carrasco, L., X. Giam, M. Papes, and K.S. Sheldon. 2019. Metrics of lidar-derived 3D vegetation structure reveal contrasting effects of horizontal and vertical forest heterogeneity on bird species richness. Remote Sensing 11:743. https://doi.org/10.3390/rs11070743
Dubayah, R.O., J. Armston, J.R. Kellner, L. Duncanson, S.P. Healey, P.L. Patterson, S. Hancock, H. Tang, J. Bruening, M.A. Hofton, J.B. Blair, and S.B. Luthcke. 2022. GEDI L4A Footprint Level Aboveground Biomass Density, Version 2.1. ORNL DAAC, Oak Ridge, Tennessee, USA. https://doi.org/10.3334/ORNLDAAC/2056
Dubayah, R.O., J. Armston, S.P. Healey, Z. Yang, P.L. Patterson, S. Saarela, G. Stahl, L. Duncanson, J.R. Kellner, J. Bruening, and A. Pascual. 2023. GEDI L4B Gridded Aboveground Biomass Density, Version 2.1. ORNL Distributed Active Archive Center. https://doi.org/10.3334/ORNLDAAC/2299
Dubayah, R., M. Hofton, J. Blair, J. Armston, H. Tang, and S. Luthcke. 2021. GEDI L2A Elevation and Height Metrics Data Global Footprint Level V002. NASA EOSDIS Land Processes Distributed Active Archive Center. https://doi.org/10.5067/GEDI/GEDI02_A.002
Dubayah, R.O., S.B. Luthcke, T.J. Sabaka, J.B. Nicholas, S. Preaux, and M.A. Hofton. 2021. GEDI L3 Gridded Land Surface Metrics, Version 2. ORNL Distributed Active Archive Center. https://doi.org/10.3334/ORNLDAAC/1952
Dubayah, R., H. Tang, J. Armston, S. Luthcke, M. Hofton, and J. Blair. 2021. GEDI L2B Canopy Cover and Vertical Profile Metrics Data Global Footprint Level V002. NASA EOSDIS Land Processes Distributed Active Archive Center. https://doi.org/10.5067/GEDI/GEDI02_B.002
Efron, B., and R.J. Tibshirani. 1994. An Introduction to the Bootstrap. Chapman and Hall/CRC; New York. https://doi.org/10.1201/9780429246593
Evans, J.S., M.A. Murphy. 2023. spatialEco. R package version 2.0-2. https://github.com/jeffreyevans/spatialEco
GDAL/OGR contributors. 2024. GDAL/OGR Geospatial Data Abstraction software Library. Open Source Geospatial Foundation. https://gdal.org
Lang, N., W. Jetz, K. Schindler, and J.D. Wegner. 2023. A high-resolution canopy height model of the Earth. Nature Ecology & Evolution 7:1778–1789. https://doi.org/10.1038/s41559-023-02206-6
Melendy, L., S. Hagen, F.B. Sullivan, T. Pearson, S.M. Walker, P. Ellis, Kustiyo, K.A. Sambodo, O. Roswintiarti, M. Hanson, A.W. Klassen, M.W. Palace, B.H. Braswell, G.M. Delgado, S.S. Saatchi, And A. Ferraz. 2017. CMS: LiDAR-derived Canopy Height, Elevation for Sites in Kalimantan, Indonesia, 2014. ORNL Distributed Active Archive Center. https://doi.org/10.3334/ORNLDAAC/1540
McRoberts, R.E., E. Næsset, Z. Hou, G. Ståhl, S. Saarela, J. Esteban, D. Travaglini, J. Mohammadi, and G. Chirici. 2023. How many bootstrap replications are necessary for estimating remote sensing-assisted, model-based standard errors? Remote Sensing of Environment 288:113455. https://doi.org/10.1016/j.rse.2023.113455
NEON. 2021. Discrete return LiDAR point cloud (DP1.30003.001). RELEASE-2022 (available at: https://data.neonscience.org ) (Accessed 16 July 2022)
Potapov, P., X. Li, A. Hernandez-Serna, A. Tyukavina, M.C. Hansen, A. Kommareddy, A. Pickens, S. Turubanova, H. Tang, C.E. Silva, J. Armston, R. Dubayah, J.B. Blair, and M. Hofton. 2021. Mapping global forest canopy height through integration of GEDI and Landsat data. Remote Sensing of Environment 253:112165. https://doi.org/10.1016/j.rse.2020.112165
R Core Team 2021 R: A language and environment for statistical computing R Foundation for Statistical Computing (available at: www.R-project.org).
Schlund, M., S. Erasmi, and A. Knohl. 2023. Rasters for ALS metrics at 10m resolution 2022. GRO.data, V1. https://doi.org/10.25625/39VQPW.
Swinfield, T., Milodowski, D., Jucker, T., Michele, D., & Coomes, D. (2020). LiDAR canopy structure 2014. Zenodo. https://doi.org/10.5281/zenodo.4020697
Torresani, M., D. Rocchini, A. Alberti, V. Moudrý, M. Heym, E. Thouverai, P. Kacic, and E. Tomelleri. 2023. LiDAR GEDI derived tree canopy height heterogeneity reveals patterns of biodiversity in forest ecosystems. Ecological Informatics 76:102082. https://doi.org/10.1016/j.ecoinf.2023.102082