Documentation Revision Date: 2024-10-02
Dataset Version: 1
Summary
There are 40 data files in cloud optimized GeoTIFF format with this dataset.
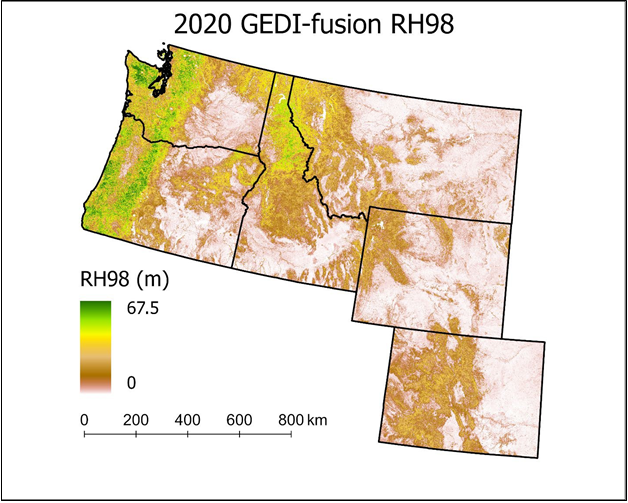
Figure 1. The browse image displays the 2020 GEDI-fusion relative height 98 (RH98) predicted map product which represents a canopy height measure across the six-state study area at a 30-m spatial resolution.
Citation
Vogeler, J., P.A. Fekety, and K. Vierling. 2023. Gridded GEDI-Fusion Forest Structure Metrics across Six Western US States, 2016-2020. ORNL DAAC, Oak Ridge, Tennessee, USA. https://doi.org/10.3334/ORNLDAAC/2236
Table of Contents
- Dataset Overview
- Data Characteristics
- Application and Derivation
- Quality Assessment
- Data Acquisition, Materials, and Methods
- Data Access
- References
Dataset Overview
This dataset provides eight GEDI forest structure metrics relevant to wildlife habitat modeling and biodiversity assessments at 30-m resolutions across Washington, Oregon, Idaho, Montana, Wyoming, and Colorado. The data were derived using random forest modeling and prediction frameworks. The models created were also hindcasted using 2019 and 2020 GEDI footprints back to 2016 on annual time steps leveraging continuous Landsat spectral and disturbance information, Sentinel-1 backscatter metrics and ratios, topographic information, and bioclimatic variables. Machine learning data fusion approaches were used to scale-up structure information provided by the novel space-borne Global Ecosystems Dynamics Investigation (GEDI) waveform lidar sensor to continuous extents using additional satellite-based continuous earth observation data. GEDI provides a consistent sample of forest structure information at 25-m diameter footprints at near-global extents, providing a valuable source of reference information to drive continuous mapping efforts.
Modeling efforts were focused on several GEDI-derived height, cover, foliage height diversity, and summarized plant area density profile metrics corresponding to important habitat structure components for a variety of wildlife species. GEDI level 2A relative height (RH) metrics represent the height at which a defined percentage of GEDI waveform energy is contained. For instance, RH98 corresponds to the height at which 98% of the waveform energy is captured - comparable to a canopy height measure. RH50 and RH75 were included within the modeling efforts to test their utility for wildlife modeling in future applied research efforts.
Among the GEDI level 2B metrics, two commonly used forest measures in wildlife habitat modeling were selected, fractional canopy cover (COVER) and foliage height diversity (FHD). Among the Level 2B plant area vegetation density (PAVD) profile metrics, the lowest single profile available through the GEDI waveform metrics was chosen that represents the proportion of vegetation within the 5-10 m strata (PAVD 5-10 m), as well as summarizing plant area proportions above 20 m (PAVD >20 m) and 40 m (PAVD >40 m) to represent the presence of a mature upper canopy within different forest types.
Related Publication
Vogeler, J.C., P.A. Fekety, L. Elliott, N.C. Swayze, S.K. Filippelli, B. Barry, J.D. Holbrook, and K.T. Vierling. 2023. Evaluating GEDI data fusions for continuous characterizations of forest wildlife habitat. Frontiers in Remote Sensing 4:1196554. https://doi.org/10.3389/frsen.2023.1196554
Acknowledgement
This work was funded by the NASA GEDI Competed Science Team 2020 (grant: 80NSSC21K0192).
Data Characteristics
Spatial Coverage: Washington, Oregon, Idaho, Montana, Wyoming, Colorado
Spatial Resolution: 30 m
Temporal Coverage: 2016 - 2020
Temporal Resolution: Annual (growing season)
Study Area: Latitude and longitude are given in decimal degrees.
| Study Area | Westernmost Longitude | Easternmost Longitude | Northernmost Latitude | Southernmost Latitude |
|---|---|---|---|---|
| Global land | -127.1130 | -101.7276 | 51.4317 | 34.3829 |
Data File Information
There are 40 data files with this dataset in cloud optimized GeoTIFF (.tif) format. Each of the eight GEDI-fusion metrics have an associated Geotiff for each year of the study period (2016-2020).
The files are named gedifusion_<metric>_<YYYY>.tif , where
- <YYYY> = year, 2016-2020
- <metric> = abbreviation of one of the eight GEDI fusion metrics listed in Table 1.
Example file name: gedifusion_pavd20m_2016.tif
GeoTIFF properties
- Coordinate system: CONUS Albers, NAD83 (EPSG: 5070)
- Spatial resolution: 30 m
- Bands: 1
- Nodata value: –3.40282306073709653e+38
Table 1. Variables in the data files.
| Vegetation metric | File name Abbreviation | Units | Description |
|---|---|---|---|
| Canopy cover | cover | 1 | Fractional canopy cover (proportion) |
| Foliage height diversity | fhd | 1 | Foliage height diversity (a unitless index) |
| Plant area vegetation density 5 to 10 m strata | pavd5to10m | 1 | Plant area vegetation density (PAVD); the proportion of vegetation within the 5-10 m stratum above ground surface (PAVD 5-10 m) |
| Plant area vegetation density greater than 20 m stratum | pavd20m | 1 | The proportion of vegetation density (PAVD) greater than 20 m above ground surface, chosen to represent the presence of a mature upper canopy within different forest types |
| Plant area vegetation density greater than 40 m stratum | pavd40m | 1 | The proportion of vegetation density (PAVD) greater than 40 m above ground surface, chosen to represent the presence of a mature upper canopy within different forest types |
| Relative height 50 | rh50 | m | Corresponds to the height at which 50% of the waveform energy is captured - comparable to a canopy height measure |
| Relative height 75 | rh75 | m | Corresponds to the height at which 75% of the waveform energy is captured - comparable to a canopy height measure |
| Relative height 98 | rh98 | m | Corresponds to the height at which 98% of the waveform energy is captured - comparable to a canopy height measure |
Application and Derivation
Spatially continuous characterizations of forest structure are critical for modeling wildlife habitat as well as for assessing trade-offs with additional ecosystem services. To overcome the spatial and temporal limitations of airborne lidar data for studying wide-ranging animals and for monitoring wildlife habitat through time, novel sampling data sources, including the space-borne Global Ecosystem Dynamics Investigation (GEDI) lidar instrument, may be incorporated within data fusion frameworks to scale up satellite-based samples of forest structure across continuous spatial extents. Project results show promise for the use of remote sensing data fusions for scaling up GEDI structure metrics of value for habitat modeling and other applications across broad continuous extents.
This data fusion approach resulted in moderate to high model performances and conducted independent map validations (see quality assessment section) across a variety of forest types, elevations, and climatic gradients.
Data fusion frameworks that are based on freely available public data, such as those from the GEDI, Landsat, and Sentinel-1 programs, support a wide variety of conservation and management applications across regions where financial resources for acquiring imagery may be limited or which lack forest inventory programs. When publicly available data are available through time, there are opportunities to hindcast spatial prediction models of forest variables in order to monitor sources of change in habitat as well as better match the timing of wildlife data collections.
Quality Assessment
Model Assessments
All model accuracies and errors were assessed using a withheld set of testing GEDI footprints (see methods section for more details). Our random forest models predicting GEDI structure metrics from continuous satellite remote sensing data sources had moderate-high model performance for the majority of the GEDI metrics (Table 2). The highest performance was observed for RH98 (R2 = 0.76, RMSE = 5.46 m) and FHD (R2 = 0.74, RMSE = 0.39). Lower model performances were observed for the PAVD metrics, with R2 values ranging from 0.36 (PAVD5-10m) to 0.58 (PAVD>20), and RMSE values ranging from 0.03 (PAVD>40m) to 0.06 (PAVD>20m).
Table 2. GEDI-fusion model assessment. All accuracy and error statistics were calculated using a withheld testing set of ~140,000 GEDI footprints.
| Vegetation metric | Abbreviation | R2 | RMSE | Bias |
|---|---|---|---|---|
| Relative height 98 | RH98 | 0.757 | 5.445 | 0.130 |
| Relative height 75 | RH75 | 0.707 | 4.238 | 0.098 |
| Relative height 50 | RH50 | 0.651 | 3.369 | 0.078 |
| Foliage height diversity | FHD | 0.739 | 0.392 | -0.005 |
| Canopy cover | COVER | 0.684 | 0.146 | 0.004 |
| Plant area vegetation density 5 to 10 m strata | PAVD5-10m | 0.363 | 0.051 | 0.002 |
| Plant area vegetation density greater than 20 m stratum | PAVD>20m | 0.580 | 0.058 | 0.001 |
| Plant area vegetation density greater than 40 m stratum | PAVD>40m | 0.447 | 0.034 | 0.001 |
GEDI Footprint-ALS Comparisons
GEDI-fusion frameworks assume that the information provided by the GEDI footprints are accurate representations of forest structure for serving as modeling reference data. To test this assumption and to inform sources of error in the GEDI-fusion maps, footprint estimates of focal metrics were compared to those from airborne laser scanning (ALS) samples. ALS data may be limited in spatial extents and temporal coverage, but the sources of errors and vertical/horizontal accuracies are well established and can serve as a baseline for comparisons with GEDI-derived forest measures and for comparing patterns in spatiotemporal biases between GEDI-fusion maps. A set of ALS collections were identified for the years of our GEDI footprint samples (2019 and 2020) that represent the forest-dominated ecoregions of the study area based on the EPA Level III Ecoregions. The sample ALS collections also captured a wide range of forest structure variability and disturbance patterns.
Comparisons between ALS simulated (RH and FHD metrics) or direct discrete ALS measures (COVER) to those from GEDI footprints showed variability in accuracies across the GEDI metrics (Table 3). The highest comparison accuracies were observed for RH98 (R2 = 0.74, RMSE = 6.83 m) and the lowest accuracies for COVER (R2 = 0.44, RMSE = 0.27). The remaining height metrics of RH75 and RH50 along with the FHD metric had comparable accuracies, with R2 values of 0.70, 0.66, and 0.61, respectively. The majority of the GEDI-fusion metrics had a negative bias with the exception of RH98, which means that the GEDI footprints tended to underestimate values compared to ALS measures. The simulated FHD values exhibited a systematic bias in that they had a higher range of values to those from the GEDI footprints (bias = -2.107), but still exhibited a good validation comparison with the actual GEDI footprint values. The lower model performances observed between the GEDI and direct ALS cover measures may be influenced by a multitude of factors; these include the different representations of cover within waveform vs. discrete lidar measures of cover, geolocation errors within the GEDI footprints, or issues with the ground finding algorithm within the GEDI footprint influencing the resulting cover estimate.
Table 3. Comparison statistics for GEDI Level 2A and 2B footprint metrics and ALS validation metrics for a sample of ALS units across our study area. Simulated waveform metrics from ALS were used for all comparisons except for COVER, which was produced directly from discrete ALS data (proportion of first returns above 2 m). Validation results are not included for PAVD metrics because those metrics are not available as outputs from the GEDI simulator or directly from discrete ALS. RMSE units are in meters for relative height (RH) metrics and proportions for COVER. FHD is unitless.
| GEDI footprint metric | R2 | RMSE | Bias |
|---|---|---|---|
| RH98 | 0.735 | 6.831 | 0.782 |
| RH75 | 0.698 | 5.629 | -0.119 |
| RH50 | 0.664 | 4.551 | -0.256 |
| FHD | 0.608 | 2.143 | -2.107 |
| COVER | 0.436 | 0.271 | -0.142 |
Structure Map Validations
In addition to comparing model performance through the withheld set of independent GEDI footprints, biases were evaluated within the predicted maps and temporal transferability of models. The sample set of ALS units was leveraged for years of model creation (2019-2020) and a set of ALS collections from 2016-2018 to represent years of model hindcasting to evaluate differences in map accuracies when models were applied to years outside of the model training data.
Map-level validations show variability in map performance across the GEDI-fusion metrics, although all had moderate-high predictive performance (R2 = 0.59-0.75). Map accuracies and errors were comparable between maps within years of model creation to those representing hindcasted years (Table 4). Among the GEDI-fusion metrics, FHD had the best map accuracy (R2 = 0.745), although it still exhibited the systematic bias observed within the footprint level comparisons. RH50 had the lowest map accuracy with an R2 = 0.59 and RMSE = 4.59 m. Map predictions underestimated RH50 compared to simulated values, particularly among higher RH50 simulated heights. The order of validation performance rankings of the GEDI-fusion metrics was different between the footprint- and map-level validations, but the general range of accuracies and errors and moderate-high performance was consistent between scales of analyses.
Table 4. GEDI-fusion gridded predicted map validation with GEDI-simulator ALS (or direct discrete ALS for COVER) sample units for maps created with the combined predictor model and the Landsat/topographic/bioclimatic model. Validation results are not included for PAVD metrics because those metrics are not available as outputs from the GEDI simulator used to simulate comparable waveform metrics from the ALS sample units, or through direct discrete ALS measures.
| GEDI Fusion Map Metric | R2 | RMSE | Bias | |
|---|---|---|---|---|
| Modeling Map Years (2019-2020) | RH98 | 0.673 | 6.996 | 1.109 |
| RH75 | 0.633 | 5.675 | 0.710 | |
| RH50 | 0.591 | 4.589 | 0.384 | |
| FHD | 0.745 | 2.197 | -2.173 | |
| COVER | 0.681 | 0.235 | -0.144 | |
| Hindcasting Map Years (2016-2018) | RH98 | 0.690 | 6.296 | 0.618 |
| RH75 | 0.650 | 5.171 | 0.563 | |
| RH50 | 0.599 | 4.329 | 0.384 | |
| FHD | 0.719 | 2.198 | -2.176 | |
| COVER | 0.653 | 0.206 | -0.126 |
Data Acquisition, Materials, and Methods
The rGEDI package was leveraged (Silva et al., 2020) and implemented in the R Statistical Software (R Core Team, 2021) to download and filter GEDI version 2 footprint data across our study area. The target GEDI footprints were restricted within a date range of June 6 - September 30 for both 2019 and 2020 to limit any bias in canopy cover and vegetation density profiles in mixed or deciduous forests outside of the primary growing season. The summer season GEDI shots were further filtered with a series of conditional arguments to retain only the highest quality observations to serve as model reference and testing data. While the rich spatial density of GEDI footprints provides value for a wide suite of applications including direct quantification and monitoring of forest patterns across broad extents, for purposes in leveraging GEDI footprints as a model reference source, GEDI footprints were spatially thinned to balance computational efficiency with maximizing model performance. The resulting spatially subsetted dataset consisted of 99,766 observations for 2019 and 100,003 observations for 2020. See Vogeler et al. (2023) for additional GEDI filtering and pre-processing steps.
Google Earth Engine (GEE) was used to generate a suite of 31 active and passive remote sensing predictor layers for upscaling GEDI forest structure metrics to a continuous 30-m resolution grid and to apply models at annual time steps from 2016-2020. All dynamic predictors (e.g., Sentinel-1 and Landsat) were summarized for the summer growing season to match the temporal window of the GEDI data. From median summer composites of the Sentinel- 1 C-band Synthetic Aperture Radar (SAR) dataset, the vertical-vertical (VV) and vertical-horizontal (VH) polarizations were compiled along with several ratios derived from the median VV and VH data. For the Landsat spectral predictors, medoid image composites were prepared for the annual summer seasons for the full Landsat archive (1984-2021), from which annual spectral indices were then calculated.
When forest attribute models are created for a particular time period and then applied across a longer time period, using a temporal segmentation fitting algorithm can aid in producing more stabilized temporal representations of the modeled attribute within predicted maps. The algorithm, LandTrendr in GEE (Kennedy et al., 2018) was used to calculate vertices within each spectral index and the original bands to produce annual “fitted” values for all Landsat predictors. In addition to the Landsat and Sentinel-1 predictor sets, disturbance histories derived from the United States Forest Service Landscape Change Monitoring System (LCMS) dataset (Housman et al., 2022) were incorporated along with topographic and bioclimatic information. See Vogeler et al. (2023) for additional details on the metrics and pre-processing of the predictor data sources.
All predictors were either aggregated or resampled to a common 30-m grid and exported from GEE for local modeling and predictive mapping using a EPSG:5070 projection within analysis ready dataset tiles. The spatially filtered GEDI locations for each year were buffered by 12.5 m to generate polygons representative of the 25-m diameter of the GEDI footprints. All predictors were extracted using an area weighted mean pixel value from all pixels intersecting a footprint’s polygon, and for temporally dynamic predictors the year used for extraction corresponded to that of the GEDI footprint’s acquisition year.
An initial evaluation of random forest regression was completed with progressively larger training samples to identify the number of training samples at which model performance began to stabilize for a sample set of GEDI metrics (i.e., a learning curve), and this number of training and testing samples was used for subsequent model development and evaluation. Model performance stabilized at approximately 60,000 training footprints in initial model testing, which was the sample size used for subsequent model development along with a withheld set of approximately 140,000 footprints for model testing. Within each single-source predictor set (e.g., Landsat, topography), tests for highly correlated variables were performed using a correlation threshold of 0.95. Only those not highly correlated were retained within the model. The final model was applied to the predictor layers to produce 30-m resolution maps of the GEDI metrics across the study area and on annual time steps from 2016-2020. As a final post-processing step, an open-water mask was developed using the Global Surface Water Layer v1.4 within GEE (Pekel et al., 2016) and applied to all final maps to minimize false vegetation structure measures as a result of the GEDI filtering approaches that removed all water GEDI points.
See Section 4 for all model performance and map validation approaches and results.
Data Access
These data are available through the Oak Ridge National Laboratory (ORNL) Distributed Active Archive Center (DAAC).
Gridded GEDI-Fusion Forest Structure Metrics across Six Western US States, 2016-2020
Contact for Data Center Access Information:
- E-mail: uso@daac.ornl.gov
- Telephone: +1 (865) 241-3952
References
Hancock, S. 2023. rGEDI Simulator software and documentation. https://bitbucket.org/StevenHancock/gedisimulator/src/master/ [Last Accessed February 7, 2023].
Hancock, S., J. Armston, M. Hofton, X. Sun, H. Tang, L. Duncanson, J.R. Kellner, and R. Dubayah. 2019. The GEDI Simulator: A large-footprint waveform Lidar simulator for calibration and validation of spaceborne missions. Earth and Space Science 6:294–310. https://doi.org/10.1029/2018EA000506
Kennedy, R.E., Z.Yang, N. Gorelick, J. Braaten, L. Cavalcante, W.B. Cohen, and S. Healey. 2018. Implementation of the LandTrendr Algorithm on Google Earth Engine. Remote Sensing 10:691. https://doi.org/10.3390/rs10050691
McGaughey, R.J. 2015. FUSION/LDV: Software for LIDAR data analysis and visualization. v.3.3. Washington, DC; USDA Forest Service. https://forsys.sefs.uw.edu/fusion/fusionlatest.html
Pekel, J.-F., A. Cottam, N. Gorelick, and A.S. Belward. 2016. High-resolution mapping of global surface water and its long-term changes. Nature, 540:418-422. https://doi.org/10.1038/nature20584
R Core Team. 2021. R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria. https://www.r-project.org/
Silva, C., C. Hamamura, R. Valbuena, S. Hancock, A. Cardil, E. Broadbent, D. Almeida, C. Silva-Junior, and C. Klauberg, C. 2020. rGEDI: An R package for NASA’s Global Ecosystem Dynamics Investigation (GEDI) data visualizing and processing. https://rgedi.r-forge.r-project.org/
Vogeler, J.C., P.A. Fekety, L. Elliott, N.C. Swayze, S.K. Filippelli, B. Barry, J.D. Holbrook, and K.T. Vierling. 2023. Evaluating GEDI data fusions for continuous characterizations of forest wildlife habitat. Frontiers in Remote Sensing 4:1196554. https://doi.org/10.3389/frsen.2023.1196554