Documentation Revision Date: 2024-06-20
Dataset Version: 4
Summary
NASA is requiring increasing compliance to open-source science approaches under its Science Information Policy SPD-41a, which states "that scientific information produced from SMD-funded scientific activities be made publicly available to the extent legally permitted". The policy goes on to say that scientific information includes both data and software. This SISTER collection is novel in that it archives not just the data products output by the SISTER workflow, but also the software and product generation environment used to produce them.
This dataset holds a total of 235 files: 8 OGC Application Packages as gzip compressed tar archives (*.tar.gz), 8 Common Workflow Language files in text format (*.cwl), 1 process file in YAML format (*.yml), 1 Jupyter notebook (*.ipynb), 2 files in comma separated values format (*.csv), 1 file of Python code (*.py), 24 binary files in ENVI format (*.bin), 24 ENVI header files (*.hdr), 15 cloud optimized GeoTIFFs (*.tif), and 107 JSON files(*.json, *.met.json).
Citation
Townsend, P., M.M. Gierach, H. Hua, S. Shah, W. Olson-Duvall, A.M. Chlus, C. Ade, O. Kwoun, M.J. Lucas, N. Malarout, D.F. Moroni, S. Neely, J.K. Pon, and D. Yu. 2024. SISTER: Experimental Workflows, Product Generation Environment, and Sample Data, V004. ORNL DAAC, Oak Ridge, Tennessee, USA. https://doi.org/10.3334/ORNLDAAC/2335
Table of Contents
- Dataset Overview
- Data Characteristics
- Application and Derivation
- Quality Assessment
- Data Acquisition, Materials, and Methods
- Data Access
- References
- Dataset Revisions
Dataset Overview
DISCLAIMER: This collection contains experimental workflows, experimental community algorithms, and experimental sample data products to demonstrate the capabilities of an end-to-end processing system. The experimental sample data products provided have not been fully validated and are not intended for scientific use. The community algorithms provided are placeholders which can be replaced by any user’s algorithms for their own science and application interests. These algorithms should not in any capacity be considered the algorithms that will be implemented in the upcoming Surface Biology and Geology mission.
This collection contains EXPERIMENTAL workflows and sample data products generated by NASA's Space-based Imaging Spectroscopy and Thermal pathfindER (SISTER) project. The SISTER project created prototype workflows and algorithms to support NASA's future Earth System Observatory's Surface Biology and Geology (SBG) mission. These workflows are specific to visible shortwave infrared (VSWIR) and do not include thermal infrared (TIR). SISTER focused on developing a data system that is open, portable, scalable, standards compliant, and reproducible.
The included files document the algorithms developed in this project and permit the user to reproduce the overall workflow or execute specific algorithms. SISTER adopted standards at various levels of production in order to make the system as open, interoperable, and reproducible as possible. These standards include:
- The Common Workflow Language (CWL) for workflow orchestration of the algorithm steps in the data production.
- Open Geospatial Consortium (OGC) Application Packages for packaging algorithms (referred to as PGEs - Product Generation Executive) that generate the data products
- The SpatioTemporal Asset Catalog (STAC) specification for describing data products used as inputs by the PGEs and generated as outputs by the PGEs.
This dataset includes: (a) the CWL process file and a Jupyter Notebook that run the entire SISTER workflow capable of generating experimental sample data products spanning terrestrial ecosystems, inland and coastal aquatic ecosystems, and snow, (b) the archived algorithm steps (as OGC Application Packages) used to generate products at each step of the workflow, (c) a small number of experimental sample data products produced by the workflow which are based on the Airborne Visible/Infrared Imaging Spectrometer-Classic (AVIRIS or AVIRIS-CL) instrument, and (d) instructions for reproducing the sample products included in this dataset.
SISTER Workflow and CWL
One of SISTER’s objectives is to prototype an SBG-like workflow that generates experimental SBG-like VSWIR data products from existing datasets over a variety of science domains including aquatic, terrestrial, and snow and ice. SISTER has adopted CWL as its framework for workflow orchestration. CWL is an open standard for creating complex data analysis workflow chains that are portable enough to run on a single laptop or on a high-performance cluster. More detail and getting started with CWL can be found at https://www.commonwl.org/.
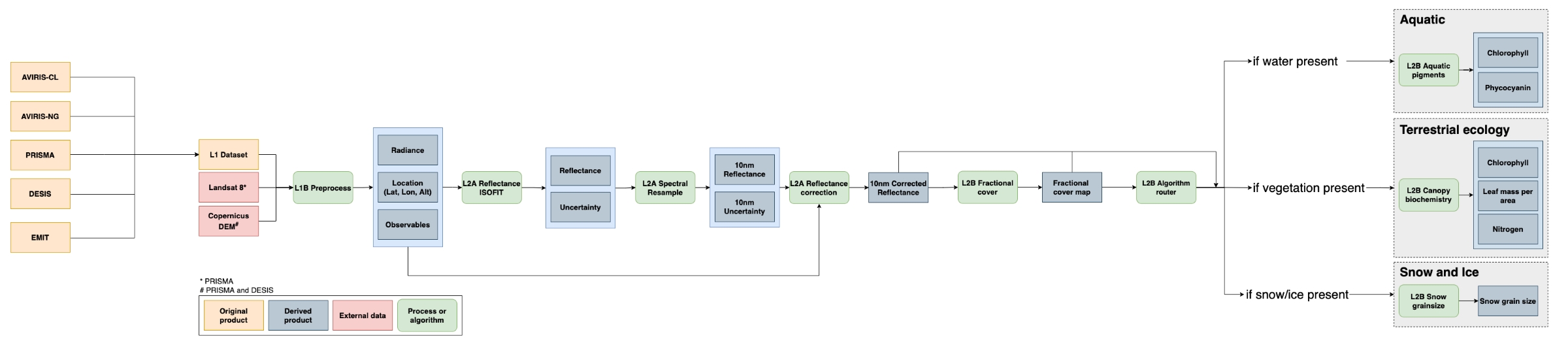
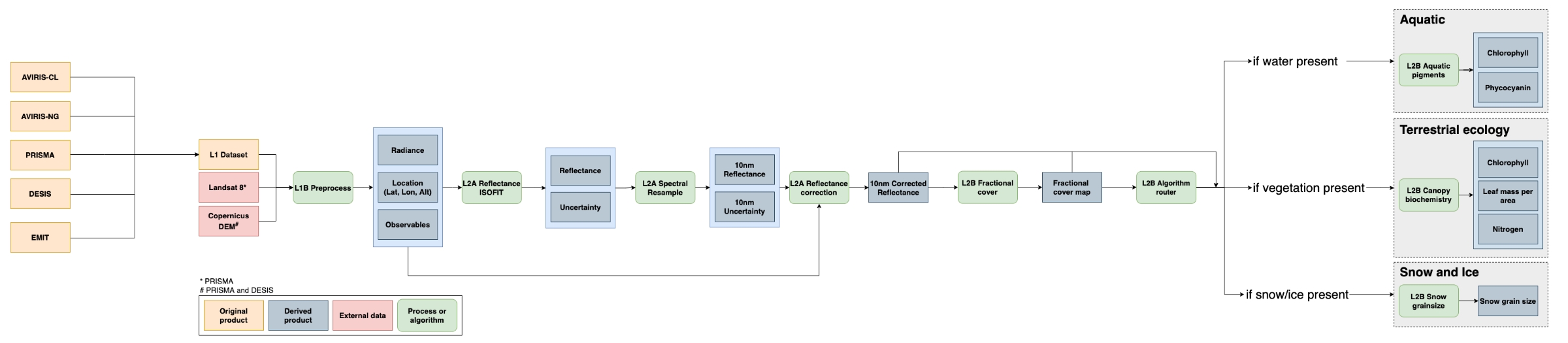
At a high level, the SISTER workflow (Figures 1, 3) starts by ingesting calibrated radiance products from a VSWIR instrument, which are spatially resampled to 30 meters, atmospherically corrected, and spectrally resampled to 10 nanometers in order to create a notional SBG-like reflectance data cube. From there, the reflectance is corrected for topography, bi-directional reflectance distribution function (BRDF) effects, and glint, after which fractional cover is estimated. Depending on the elements found in the scene, the data are then used to generate measures of aquatic pigments, vegetative traits, or snow-and-ice data products. More details can be found in Section 5 below.
OGC Application Packages and Composite Release ID
The SBG landscape of community science algorithms is large, so SISTER adopted an open standard for implementing its example algorithms. OGC Application Packages were used for algorithm packaging. OGC is the Open Geospatial Consortium, which defines standards in order to “make location information FAIR – Findable, Accessible, Interoperable, and Reusable” (https://www.ogc.org/).
OGC Application Packages consist of an executable container (like a Docker container) and a process.cwl file that describes the container’s inputs and tells it how to run (Figure 2). The container is built from code contained in a repository (e.g., Github.com) and includes all the packages and libraries needed for the code to run, making the OGC Application Package entirely portable. Any system that is running CWL can also run an OGC Application Package. In Figure 2, the “Container Registry” can be a remote registry, or a local instance on the user’s laptop.
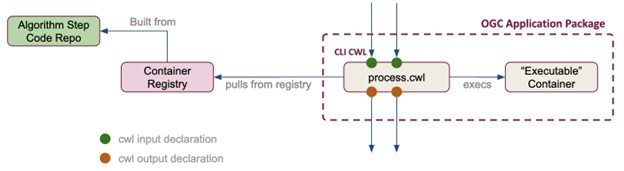
Figure 2. An OGC application package includes the process.cwl file and an executable container. The application package may interact with a local or remote registry and repository.
In the case of SISTER, all the algorithms in the SISTER workflow (e.g. Corrected Reflectance, Fractional Cover, etc) have been built into OGC Application Packages included in this dataset and archived using Zenodo.
This version of SISTER has been tagged with the Composite Release ID (CRID) of 004. CRID 004 refers to the entirety of the system used to generate the experimental SISTER data products, including the overall CWL workflow and the specific versions and DOIs of each PGE.
Table 1. Components of Composite Release ID (CRID) 004 with associated repositories in GitHub and Zenodo.
The DOIs point to Zenodo repositories that hold the source code, CWL process file (*.cwl) and a Docker image for creating an executable container (*.tar.gz) for each PGE. The CWL and container image files are the same as provided in this collection.
Data Products and AVIRIS Classic
SISTER endeavored to prototype experimental sample data products that are SBG-like in form, although these products were not validated and are not intended for scientific use. Any products generated by this system are only intended to be used to prototype what future SBG products may look like in terms of file format, naming convention, and metadata.
In particular, SISTER adopted the STAC specification as a way of standardizing the inputs and outputs of each PGE. STAC is an open standard that provides a common language for describing and cataloging spatiotemporal assets (https://stacspec.org). Currently, SISTER produces data products in both ENVI and cloud optimized GeoTIFF (COG) formats, and every published product contains its own corresponding GeoJSON metadata file (*.json). This STAC compliant file provides a standard way for describing each product and also makes the SISTER data products compatible with a wide range of community developed tools in the STAC ecosystem (https://stacindex.org/ecosystem).
Since SBG standard products do not yet exist, SISTER relied on analog imaging spectrometers for input data. This workflow has been tested with data from the Airborne Visible/Infrared Imaging Spectrometer-Classic (AVIRIS or AVIRIS-CL) instrument, the Airborne Visible/Infrared Imaging Spectrometer-Next Generation (AVIRIS-NG) instrument, the PRecursore IperSpettrale della Missione Applicativa (PRISMA) satellite, the DLR Earth Sensing Imaging Spectrometer (DESIS) instrument, and the Earth Surface Mineral Dust Source Investigation (EMIT) instrument. However, only a handful of AVIRIS-Classic scenes have been selected to demonstrate this workflow for this dataset. Radiance data from three AVIRIS-Classic scenes (Table 2) were used to generate the SISTER data products provided in this collection (Table 3).
Table 2. AVIRIS-Classic radiance datasets used as input for SISTER Product Generation Executives (PGE) algorithms in the sample workflows.
| PGE | Download URL for Radiance files |
|---|---|
| Aquatic Pigments | https://popo.jpl.nasa.gov/avcl/y18_data/f180126t01p00r08.tar.gz |
| Snow Grain Size | https://popo.jpl.nasa.gov/avcl/y11_data/f110513t01p00r03.tar.gz |
| Vegetative Traits | https://popo.jpl.nasa.gov/avcl/y13_data/f130612t01p00r10.tar.gz |
User Note: The AVIRIS-Classic radiance data is also available from the ORNL DAAC's AVIRIS-Classic L1B Calibrated Radiance Facility Instrument collection (Green et al., 2023). File information with download URLs for the ORNL DAAC are listed in the file SISTER_Workflow_AVCL_files.csv. The "CMR_query" field provides a link for obtaining the most up-to-date metadata and access information for each file in JSON format. If the user obtains radiance files from the ORNL DAAC, then the code in the Jupyter notebook (sister_cwl_workflow.ipynb, used to run the entire workflow) must be modified to reflect the change in file locations.
Table 3. Output products from SISTER Product Generation Executives (PGE) algorithms with file naming components (<sensor>_<processing level>_<product>).
| Product Description | File name component |
|---|---|
| Preprocessed Radiance, Observation, and Location | AVCL_L1B_RDN |
| Surface Reflectance and Uncertainty | AVCL_L2A_RFL |
| Resampled Surface Reflectance and Uncertainty | AVCL_L2A_RSRFL |
| Topo, BRDF, Glint Corrected Surface Reflectance | AVCL_L2A_CORFL |
| Fractional Cover | AVCL_L2B_FRCOV |
| Aquatic Pigments | AVCL_L2B_AQUAPIG |
| Vegetative Traits | AVCL_L2B_VEGBIOCHEM |
| Snow Grain Size | AVCL_L2B_SNOWGRAIN |
How to Reproduce These Results
DISCLAIMER: This collection contains experimental workflows, experimental community algorithms, and experimental sample data products to demonstrate the capabilities of an end-to-end processing system. The experimental sample data products provided have not been fully validated and are not intended for scientific use. The community algorithms provided are placeholders which can be replaced by any user’s algorithms for their own science and application interests. These algorithms should not in any capacity be considered the algorithms that will be implemented in the upcoming Surface Biology and Geology (SBG) mission.
The questions of what kind of science data system (SDS) and which algorithms to use for SBG are still undecided. The final system may be a mix of cloud and on-premises processing combined with an array of community science algorithms. SISTER has not attempted to answer these questions, but instead to develop a system that is portable enough to run on any kind of platform and open and flexible enough to support any number of science algorithms. To the extent possible, every element of SISTER is open and available to researchers and developers. Here is a simple guide to reproducing the results of any of the PGEs in the SISTER workflow.
Install Dependencies
First, there are two dependencies to run a PGE on any system, CWL and Docker. Each PGE is wrapped up as an OGC Application Package, which consists of a CWL file and a Docker image for creating an executable container.
To install CWL, Python (https://www.python.org/downloads/) or Anaconda (https://www.anaconda.com/download) may be needed. Using these tools, install CWL using
pip install cwltool
or
conda install -c conda-forge cwltool
To install Docker, follow the instructions at https://docs.docker.com/engine/install/.
Get the OGC Application Package and Inputs
This example uses the spectral resampling PGE and requires the container file “ogc-app-pack-sister-resample__3.0.0.tar.gz” and the CWL process file “ogc-sister-resample.3.0.0.process.cwl” (Table 5). The “ogc-app-pack-*.tar.gz” files are Docker images that, when loaded, create the Docker container needed to execute the package (Figure 2). Download and install both files on your local system in the same directory. Once the files are downloaded, load the container image into Docker using the command:
docker load -i ogc-app-pack-sister-resample__3.0.0.tar.gz
Also, download any required input files from this collection to the same directory as the CWL file. The specific input arguments for each PGE are described in Table 4 below.
The spectral resampling PGE requires both a reflectance dataset and an uncertainty dataset as input (Table 4). Input files should be from a single flight scene (e.g., AVIRIS-CL 20110513T175417). For each "dataset" input, create a folder with the dataset name, then download the data file(s) and STAC JSON file into the folder. Each input dataset should be in its own separate folder.
For example, a reflectance dataset input would look like...
SISTER_AVCL_L2A_RFL_20110513T175417_004/EXPERIMENTAL-SISTER_AVCL_L2A_RFL_20110513T175417_004.bin SISTER_AVCL_L2A_RFL_20110513T175417_004/EXPERIMENTAL-SISTER_AVCL_L2A_RFL_20110513T175417_004.hdr SISTER_AVCL_L2A_RFL_20110513T175417_004/EXPERIMENTAL-SISTER_AVCL_L2A_RFL_20110513T175417_004.json
and the uncertainty input will be provided as...
SISTER_AVCL_L2A_RFL_20110513T175417_004_UNC/EXPERIMENTAL-SISTER_AVCL_L2A_RFL_20110513T175417_004_UNC.bin SISTER_AVCL_L2A_RFL_20110513T175417_004_UNC/EXPERIMENTAL-SISTER_AVCL_L2A_RFL_20110513T175417_004_UNC.hdr SISTER_AVCL_L2A_RFL_20110513T175417_004_UNC/EXPERIMENTAL-SISTER_AVCL_L2A_RFL_20110513T175417_004_UNC.json
Note that there may be other related reflectance files like a browse image (*.png) or run log (*.log), but only the actual data file(s) (in this case the “.bin” and “.hdr” ENVI files) and the STAC metadata file (*.json) are required.
Run the PGE
Execute the PGE using the command:
cwltool ogc-sister-resample.3.0.0.process.cwl#process --crid "004" --experimental "True" --reflectance_dataset SISTER_AVCL_L2A_RFL_20110513T175417_004 --uncertainty_dataset SISTER_AVCL_L2A_RFL_20110513T175417_004_UNC
All PGEs use this same command format to execute their containers. The generic form of this command is:
cwltool CWL_PROCESS_FILE#process --ARG1 ARG1_VALUE --ARG2 ARG2_VALUE --ARG3 ARG3_VALUE ...
For this example, the expected output files are:
EXPERIMENTAL-SISTER_AVCL_L2A_RSRFL_20110513T175417_004.bin EXPERIMENTAL-SISTER_AVCL_L2A_RSRFL_20110513T175417_004.hdr EXPERIMENTAL-SISTER_AVCL_L2A_RSRFL_20110513T175417_004.json EXPERIMENTAL-SISTER_AVCL_L2A_RSRFL_20110513T175417_004.log EXPERIMENTAL-SISTER_AVCL_L2A_RSRFL_20110513T175417_004.met.json EXPERIMENTAL-SISTER_AVCL_L2A_RSRFL_20110513T175417_004.png EXPERIMENTAL-SISTER_AVCL_L2A_RSRFL_20110513T175417_004.runconfig.json EXPERIMENTAL-SISTER_AVCL_L2A_RSRFL_20110513T175417_004_UNC.bin EXPERIMENTAL-SISTER_AVCL_L2A_RSRFL_20110513T175417_004_UNC.hdr EXPERIMENTAL-SISTER_AVCL_L2A_RSRFL_20110513T175417_004_UNC.json EXPERIMENTAL-SISTER_AVCL_L2A_RSRFL_20110513T175417_004_UNC.met.json
Output files of this PGE follow this naming convention:
(EXPERIMENTAL-)SISTER_<SENSOR>_L2A_RSRFL_<YYYYMMDDTHHMMSS>_<CRID>_<SUBPRODUCT>.<ext>
The "EXPERIMENTAL-" prefix is optional and is only added when the "--experimental" input argument is set to True. The "crid" argument determines the <CRID> portion of the output file names.
Note: The commands for running the Fractional Cover PGE differ from those described in this section. Those commands are provided after Table 4 below.
Table of Inputs
Inputs for each of the PGEs are listed in Table 4 and can be found in the “README.md” and the “algorithm_config.yaml” files included in each source code archive listed in Table 1. For example, these files for the spectral resampling PGE can be found at:
- https://github.com/sister-jpl/sister-resample/blob/3.0.0/README.md
- https://github.com/sister-jpl/sister-resample/blob/3.0.0/algorithm_config.yaml
Table 4. Inputs required for each PGE’s OGC Application Package. For a given run, the input files should be from a single flight scene.
| PGE | Input arguments | Input Files (from this collection) |
|---|---|---|
| Preprocessing | raw_dataset crid (“000”) experimental ("True") |
AVIRIS-Classic radiance files. See Table 1 for download source. |
| Reflectance/Isofit | radiance_dataset location_dataset observation_dataset segmentation_size (“50”) n_cores (“32”) crid (“000”) experimental (“True”) |
EXPERIMENTAL-SISTER_AVCL_L1B_RDN*.bin EXPERIMENTAL-SISTER_AVCL_L1B_RDN*.hdr EXPERIMENTAL-SISTER_AVCL_L1B_RDN*.json EXPERIMENTAL-SISTER_AVCL_L1B_RDN_*_LOC*.bin EXPERIMENTAL-SISTER_AVCL_L1B_RDN_*_LOC*.hdr EXPERIMENTAL-SISTER_AVCL_L1B_RDN_*_LOC*.json EXPERIMENTAL-SISTER_AVCL_L1B_RDN_*_OBS*.bin EXPERIMENTAL-SISTER_AVCL_L1B_RDN_*_OBS*.hdr EXPERIMENTAL-SISTER_AVCL_L1B_RDN_*_OBS*.json |
| Resampled Reflectance | reflectance_dataset uncertainty_dataset crid (“000”) experimental (“True”) |
EXPERIMENTAL-SISTER_AVCL_L2A_RFL_*.bin EXPERIMENTAL-SISTER_AVCL_L2A_RFL_*.hdr EXPERIMENTAL-SISTER_AVCL_L2A_RFL_*.json EXPERIMENTAL-SISTER_AVCL_L2A_RFL_*UNC.bin EXPERIMENTAL-SISTER_AVCL_L2A_RFL_*.UNC.hdr EXPERIMENTAL-SISTER_AVCL_L2A_RFL_*.UNC.json |
| Topo, BRDF, Glint Corrected Reflectance | reflectance_dataset* observation_dataset crid (“000”) experimental (“True”) |
EXPERIMENTAL-SISTER_AVCL_L2A_RSRFL_*.bin EXPERIMENTAL-SISTER_AVCL_L2A_RSRFL_*.hdr EXPERIMENTAL-SISTER_AVCL_L2A_RSRFL_*.json EXPERIMENTAL-SISTER_AVCL_L2A_RSRFL_*UNC.bin EXPERIMENTAL-SISTER_AVCL_L2A_RSRFL_*.UNC.hdr EXPERIMENTAL-SISTER_AVCL_L2A_RSRFL_*.UNC.json |
| Fractional Cover**** |
reflectance_dataset** |
EXPERIMENTAL-SISTER_AVCL_L2A_CORFL_*.bin EXPERIMENTAL-SISTER_AVCL_L2A_CORFL_*.hdr EXPERIMENTAL-SISTER_AVCL_L2A_CORFL_*.json |
| Aquatic Pigments | corrected_reflectance_dataset** fractional_cover_dataset*** crid (“000”) experimental (“True”) |
EXPERIMENTAL-SISTER_AVCL_L2A_CORFL_*.bin EXPERIMENTAL-SISTER_AVCL_L2A_CORFL_*.hdr EXPERIMENTAL-SISTER_AVCL_L2A_CORFL_*.json EXPERIMENTAL-SISTER_AVCL_L2B_FRCOV_*.tif |
| Vegetative Traits | reflectance_dataset** frcov_dataset*** veg_cover (“0.5”) crid (“000”) experimental (“True”) |
EXPERIMENTAL-SISTER_AVCL_L2A_CORFL_*.bin EXPERIMENTAL-SISTER_AVCL_L2A_CORFL_*.hdr EXPERIMENTAL-SISTER_AVCL_L2A_CORFL_*.json EXPERIMENTAL-SISTER_AVCL_L2B_FRCOV_*.tif |
| Snow Grain Size | reflectance_dataset** frcov_dataset*** snow_cover (“0.9”) crid (“000”) experimental (“True”) |
EXPERIMENTAL-SISTER_AVCL_L2A_CORFL_*.bin EXPERIMENTAL-SISTER_AVCL_L2A_CORFL_*.hdr EXPERIMENTAL-SISTER_AVCL_L2A_CORFL_*.json EXPERIMENTAL-SISTER_AVCL_L2B_FRCOV_*.tif |
*Although the input argument is "reflectance_dataset", this PGE requires the 10-nm spectrally resampled reflectance data as input.
**These inputs require the topo, BRDF, glint corrected reflectance product as input.
***These PGEs require the fractional cover data as input. Unfortunately, spelling of this input argument varies among PGEs.
****The Fractional Cover PGE cannot be executed using the method described above. See the instructions below.
Running the Fractional Cover PGE
The Fractional Cover PGE is executed using the docker run command rather than cwltool. Use these steps:
- Create a working directory (<fraccov>) that is writeable by any user (e.g., chmod -R 777 <fraccov>).
- Copy input files (Table 4) and docker image (ogc-app-pack-sister-fractional-cover__2.0.0.tar.gz) to the <fraccov> directory.
- Execute the command:
docker run -v<path to fraccov>:/data/workdir -w /data/workdir ogc-app-pack-sister-fractional-cover:2.0.0 /app/sister-fractional-cover/run.sh --n_cores 2 --refl_scale 0.4 --crid 004 --experimental true --reflectance_dataset /data/workdir/EXPERIMENTAL-SISTER_AVCL_L2A_CORFL_20130612T181031_004
A Note about Running the Entire Workflow
The instructions above describe how to reproduce individual data products from each of the PGE steps. In addition, the entire SISTER workflow is also runnable using the workflow CWL archived in this dataset. At this time, however, running the entire workflow relies on an additional dependency called MAAP (Multi-Mission Algorithm and Analysis Platform - https://www.earthdata.nasa.gov/esds/maap) which typically requires a more advanced installation as well as a system capable of running production-level operations. The PGE for estimating atmospheric correction alone will require a high-end server supporting 30+ cores. If these dependencies are met, a user will be able to reproduce the entirety of the workflow using MAAP and the Jupyter Notebook included in this dataset.
Data Characteristics
NOTE: The characteristics for these experimental prototype data were selected to be in family with SBG-VSWIR data, but do not reflect the actual characteristics of SBG-VSWIR data, which is still in formulation at this time. The SBG-TIR data was not included as part of this prototype effort.
Spatial Coverage: Selected AVIRIS-Classic flight scenes from Hawaii, California, and Colorado
Spatial Resolution: 30 m
Spectral Range and Resolution: 400 nm to 2500 nm in 10-nm intervals
Temporal Resolution: One-time estimates
Temporal Coverage: 2011-05-13 to 2018-01-26
Study Area: (All latitudes and longitudes given in decimal degrees)
| Site | Westernmost Longitude | Easternmost Longitude | Northernmost Latitude | Southernmost Latitude |
|---|---|---|---|---|
| Hawaii, California, Colorado | -158.06 | -107.96 | 39.08 | 21.20 |
Data File Information
This dataset holds a total of 235 files: 8 OGC Application Packages as gzip compressed tar archives (*.tar.gz), 8 Common Workflow Language files in text format (*.cwl), 1 process file in YAML format (*.yml), 1 Jupyter notebook (*.ipynb), 2 files in comma separated values format (*.csv), 1 file of Python code (*.py), 24 binary files in ENVI format (*.bin), 24 ENVI header files (*.hdr), 15 cloud optimized GeoTIFFs (*.tif), and 107 JSON files(*.json, *.met.json).
Workflow Files
The PGE workflows are represented by OGC Application Packages with an associated CWL process file.
Table 5. Files for OGC Application packages by SISTER workflow algorithm. The *.tar.gz files are Docker images for creating executable containers, and the *.cwl files describe the container’s inputs and processing information.
| PGE Algorithm | OGC Application Package (*.tar.gz) with Process file (*.cwl) |
|---|---|
| Radiance preprocessing | ogc-app-pack-sister-preprocess__3.0.0.tar.gz ogc-sister-preprocess.3.0.0.process.cwl |
| Reflectance / Isofit | ogc-app-pack-sister-isofit__3.0.0.tar.gz ogc-sister-isofit.3.0.0.process.cwl |
| Spectrally resampled reflectance | ogc-app-pack-sister-resample__3.0.0.tar.gz ogc-sister-resample.3.0.0.process.cwl |
| Topo, BRDF, Glint Corrected Reflectance | ogc-app-pack-sister-reflect_correct__3.0.0.tar.gz ogc-sister-reflect_correct.3.0.0.process.cwl |
| Fractional Cover | ogc-app-pack-sister-fractional-cover__2.0.0.tar.gz ogc-sister-fractional-cover.2.0.0.process.cwl |
| Aquatic Pigments | ogc-app-pack-sister-aquatic-pigments-pge__2.0.0.tar.gz ogc-sister-aquatic-pigments-pge.2.0.0.process.cwl |
| Vegetative Traits | ogc-app-pack-sister-trait_estimate__2.0.0.tar.gz ogc-sister-trait_estimate.2.0.0.process.cwl |
| Snow Grain Size | ogc-app-pack-sister-grainsize__2.0.0.tar.gz ogc-sister-grainsize.2.0.0.process.cwl |
These files may be used to execute the workflow pipeline in its entirety:
- sister-workflow-conditional.yml: Entire workflow described in CWL
- sister_cwl_workflow.ipynb: Jupyter notebook
- sister_production_5_list.csv: List of data inputs used by Jupyter notebook
- sister_router.py: Production rule (algorithm router) for L2B products
- sister_workflow_AVCL_files.csv: List of AVIRIS-Classic radiance files with download information as alternative to URLs in Table 2.
Sample Output Products
There are 214 files of sample output products generated from three AVIRIS-Classic flight lines (Tables 3 and 6). Each flight line includes:
- Preprocessed radiance, location, and observation data at 30 m spatial resolution and native spectral resolution in ENVI binary and header pairs, with an associated quicklook image of radiance and processing product generation information for experimental reproducibility. In all, there are 15 files per flight line for this product.
- Reflectance and uncertainty at 30 m spatial resolution and native spectral resolution in ENVI binary and header pairs, with an associated quicklook image of reflectance and processing product generation information for experimental reproducibility. In all, there are 11 files per flight line for this product.
- Resampled reflectance and uncertainty at 30 m spatial resolution and 10 nm spectral resolution in ENVI binary and header pairs, with an associated quicklook image of reflectance and processing product generation information for experimental reproducibility. In all, there are 11 files per flight line for this product.
- Reflectance corrected for topography, bi-directional reflectance distribution function (BRDF) effects, and glint in ENVI binary and header pairs with an associated quicklook image of reflectance as well as processing product generation information for experimental reproducibility. In all, there are 7 files per flight line for this product.
- Fractional cover of soil, vegetation, water, and snow in cloud optimized GeoTIFF format, with an associated quicklook image and processing product generation information for experimental reproducibility. In all, there are 6 files per flight line for this product.
In addition, each flight line was chosen to demonstrate a higher-level data product in a particular science area (Table 2). Depending on the content of each scene, the flight lines may also contain:
- Aquatic pigments (chlorophyll A and phycocyanin concentrations) in cloud optimized GeoTIFF format. Additional files for each flightline include a quicklook image as well as processing product generation information for experimental reproducibility and a PGE log file. In all, there are 24 files per flight line for this product.
- Vegetative traits (chlorophyll content, nitrogen concentration, leaf mass per area) in cloud optimized GeoTIFF format. Additional files for each flightline include a quicklook image as well as processing product generation information for experimental reproducibility and a Product Generation Executive (PGE) log file. In all, there are 28 files per flight line for this product.
- Snow grain size in Cloud Optimized GeoTIFF format with an associated quicklook image as well as processing product generation information for experimental reproducibility. In all, there are 12 files per flight line for this product.
File naming convention for output products:
EXPERIMENTAL-SISTER_<instrument>_<level>_<product>_<flight_id>_<ver>_<subproduct>.<ext>, where
- <instrument> = the spectroscopy instrument that provided input radiance: "AVCL" (AVIRIS-Classic)
- <level> = the NASA Earthdata Data Processing Level
- <product> = the SISTER Project data product (Table 6)
- <flight_id> = flight line identifier, <YYYYMMDD>T<hhmmss>, encoding the date and time by year (YYYY), month (MM), day (DD), and the UTC hour (hh), minute (mm), and second (ss) for the start of flight line.
- <ver> = the SISTER processing version, also known as the CRID
- <subproduct> = optional modifier for some products. See Table 6.
- <ext> = file extension indicating file format: “bin” (ENVI binary file), “hdr” (ENVI header file), “tif” (GeoTIFF), “png” (PNG image), “json” (JSON text), “log” (logging information in text format). See Table 7.
Table 6. Output product file types in ENVI and GeoTIFF formats. The units for these file types are "1" unless otherwise noted in the description.
| Product abbreviation | Subproduct | Description | File format |
|---|---|---|---|
| RDN | - | Radiance image cube ( mW cm-2 nm-1 sr-1) | ENVI: binary file (bin) with header (hdr) |
| LOC | Location: Orthocorrected pixel locations for each radiance image cube in geographic coordinates as decimal degrees in WGS-84 datum and estimated ground elevation at pixel center in m | ||
| OBS | Observational information: Parameters related to the geometry of observation and illumination for each pixel. | ||
| RFL | - | Reflectance | |
| UNC | Reflectance uncertainty | ||
| RSRFL | - | Resampled reflectance: resampled to 10-nm spectral resolution | |
| UNC | Resampled reflectance uncertainty | ||
| CORFL | - | Corrected reflectance: corrected for topography, bi-directional reflectance distribution function (BRDF) effects, and glint | |
| UNC | Corrected reflectance uncertainty | ||
| FRCOV | - | Fractional cover of soil, vegetation, water, and snow; 4 bands | GeoTIFF |
| AQUAPIG | CHL | Aquatic pigments: Chlorophyll a concentration (mg m-3) | |
| PHYCO | Aquatic pigments: Phycocyanin concentration (mg m-3) | ||
| VEGBIOCHEM | CHL | Vegetative traits: Chlorophyll concentration (µg cm-2) | |
| LMA | Vegetative traits: Leaf mass per area (g m-2) | ||
| NIT | Vegetative traits: Nitrogen concentration (mg g-1) | ||
| SNOWGRAIN | - | Snow grain size (µm) and Quality assurance mask; 2 bands |
Table 7. Additional text and image files that accompany products.
| File extension | Description |
|---|---|
| *.png | Quicklook image in PNG format |
| *_<subproduct>.json | STAC compliant metadata including sensor, start and end time, description, bounding box, product, and processing level |
| *.met.json | PGE run metadata that documents name of the algorithm and related information |
| *.runconfig.json | PGE runconfig: defines inputs for the dataset's run |
| *.log | PGE log information |
ENVI files include a binary data file (.bin) and an accompanying header file (.hdr) in text format. The ENVI header (https://www.l3harrisgeospatial.com/docs/enviheaderfiles.html) holds metadata for the binary data file, including:
- number of samples (columns), lines (rows), and bands
- band information: wavelength and full width at half maximum (fwhm)
- data type (4 = Float32, 5=Float64), interleave type, and byte order
- map info: projection and datum, coordinates for x y reference points, pixel size, and map units
- file metadata: sensor type, start and end acquisition time, and bounding box
GeoTIFF characteristics:
- Coordinate system: UTM projection, WGS84 datum; UTM zones 4N (Hawaii, EPSG 32604), 11N (California, EPSG 32611), and 12N (Colorado, EPSG 32612).
- Spatial resolution: 30 m
- Bands: varies by product, see Table 6.
- Pixel values/units: varies by product, see Table 6.
- NoData value: -9999
Application and Derivation
The NASA Earth System Observatory (ESO) is a set of Earth-focused missions, identified in the 2018 National Academies’ Decadal Survey “Thriving on Our Changing Planet: A Decadal Strategy for Earth Observation from Space,” that will provide key information to guide efforts related to climate change. Surface Biology and Geology (SBG) is one of the NASA ESO missions. SBG will provide near-global, high-spatial-resolution spectral imaging of the land surface and adjacent coastal zones to quantify Earth system processes in the areas of terrestrial and aquatic ecosystems, Earth’s surface and Interior, and hydrology. SISTER originated in support of the SBG mission to develop prototype workflows with community algorithms and generate prototype data products envisioned for SBG.
This collection is the result of the SISTER activity. As an open, portable, and reproducible system, it can represent:
- Workflow blueprint for researchers to test algorithms.
- Model of a mission science data system (SDS) that is compliant with NASA Science Information Policy SPD-41a for open-source science.
- Example file format and metadata for SBG-like data products.
The characteristics for these experimental prototype data were selected to be in family with SBG-VSWIR data, but do not reflect the actual characteristics of SBG-VSWIR data, which is still in formulation at this time. The SBG-TIR data was not included as part of this prototype effort.
The output products were not validated and are not intended for scientific use.
Quality Assessment
This collection contains experimental workflows, experimental community algorithms, and experimental sample data products to demonstrate the capabilities of an end-to-end processing system. The sample data products provided have not been fully validated and are not intended for scientific use. The community algorithms provided are placeholders that can be replaced by any user’s algorithms for their own science and application interests. These algorithms should not in any capacity be considered the algorithms that will be implemented in the upcoming SBG mission.
The output products were not validated and are not intended for scientific use.
Data Acquisition, Materials, and Methods
The input flight lines in this collection were collected by the AVIRIS Classic (AVIRIS-CL) imaging spectrometer as part of various flight campaigns in the western United States over vegetation, aquatic and snow targets. AVIRIS-CL measures radiance at approximately 10-nanometer (nm) intervals in the visible to shortwave infrared spectral range between 400 and 2500 nm. The raw AVIRIS data were processed to orthorectified calibrated radiance by the Imaging Spectroscopy Group at JPL and then ingested into the SISTER platform for further downstream processing.
Figure 3. Workflow diagram of the SISTER 004 production run (Click on image to view full-resolution version). The SISTER workflow was tested to work with inputs from multiple spectrometers; however, this dataset only includes outputs from AVIRIS-Classic.
First, images were spatially downsampled to 30-meter resolution and processed to surface reflectance using an optimal estimation atmospheric correction algorithm, ISOFIT (Thompson et al., 2018), with an open-source neural-network-based emulator for modeling radiative transfer (Brodrick et al., 2021). Next, spectral resampling to a 10-nm sampling interval was performed in a two-step calculation. Bands were first aggregated and averaged to the closest resolution to the target interval then a piecewise cubic interpolator was used to interpolate the spectra to the target wavelength spacing.
Following spectral resampling, a combination of topographic, bidirectional reflectance distribution function (BRDF) and glint correction algorithms were applied to each image to generate corrected reflectance. Topographic correction was performed using the Sun-Canopy-Sensor+C algorithm (Soenen et al., 2005), BRDF correction was performed using the FlexBRDF algorithm (Queally et al., 2022) and glint correction was performed using the method of Gao and Li (2021).
Fractional cover maps were generated from the corrected reflectance datasets using a spectral mixture analysis (Keshava and Mustard 2002). Spectral unmixing was performed using a four endmember dataset derived from the EMIT surface reflectance library (Thompson 2021a; 2021b; 2021c) to estimate fractional cover estimates of soil, vegetation, water and snow/ice. A brightness normalization was applied to minimize the impact of intraclass brightness variability on unmixing results.
Maps of aquatic phycocyanin and chlorophyll concentrations were generated from corrected reflectance and fractional cover datasets using pre-trained mixture density networks (MDN). MDN weights were derived from Pahlevan et al. (2021) for chlorophyll and O'Shea et al. (2021) for phycocyanin. Prior to model application, reflectance datasets were resampled to model wavelengths. Pigment concentrations were only estimated for pixels with water fractional cover of greater than 0.9.
Terrestrial vegetation biochemistry was estimated using partial least squares regression (PLSR) models. Permuted PLSR models for estimating chlorophyll content, nitrogen concentration, and leaf mass per area were developed using coincident NEON AOP canopy spectra, downsampled to 10 nm, and field data collected by Wang et al. (2020). Biochemical trait estimates were only calculated for pixels with greater than 50% vegetation cover. In addition to mean biochemical trait estimates, per-pixel uncertainties were calculated along with a quality assurance mask that flags pixels with trait estimates outside of the range of data used to build the model.
Snow grain size was estimated from the corrected reflectance datasets using the method of Nolin and Dozier (2000), which models grain size as a function of scaled band area centered at the 1030 nm ice absorption feature. Snow grain size was only calculated for pixels with greater than 90% snow cover. A quality assurance mask was created that flags pixels with grain size estimates outside of the range of the model (60 - 1000 microns).
Data Access
These data are available through the Oak Ridge National Laboratory (ORNL) Distributed Active Archive Center (DAAC).
SISTER: Experimental Workflows, Product Generation Environment, and Sample Data, V004
Contact for Data Center Access Information:
- E-mail: uso@daac.ornl.gov
- Telephone: +1 (865) 241-3952
References
Brodrick, P.G., D.R. Thompson, J.E. Fahlen, M.L. Eastwood, C.M. Sarture, S.R. Lundeen, W. Olson-Duvall, N. Carmon, and R.O. Green. 2021. Generalized radiative transfer emulation for imaging spectroscopy reflectance retrievals, Remote Sensing of Environment 261:112476. https://doi.org/10.1016/j.rse.2021.112476
Gao, B.C., and R.R. Li. 2021. Correction of Sunglint Effects in High Spatial Resolution Hyperspectral Imagery Using SWIR or NIR Bands and Taking Account of Spectral Variation of Refractive Index of Water. Advances in Environmental and Engineering Research 2:1-15. https://doi.org/10.21926/aeer.2103017
Green, R.O., D.R. Thompson, J.W. Boardman, J.W. Chapman, M. Eastwood, M. Helmlinger, S.R. Lundeen, and W. Olson-Duvall. 2023. AVIRIS-Classic: L1B Calibrated Radiance, Facility Instrument Collection, V1. ORNL DAAC, Oak Ridge, Tennessee, USA. https://doi.org/10.3334/ORNLDAAC/2155
Keshava, N. and J.F. Mustard. 2002. Spectral unmixing. IEEE Signal Processing Magazine 19:44-57. https://doi.org/10.1109/79.974727
Nolin, A.W. and J. Dozier. 2000. A hyperspectral method for remotely sensing the grain size of snow. Remote Sensing of Environment 74:207-216. https://doi.org/10.1016/S0034-4257(00)00111-5
O'Shea, R. E., N. Pahlevan, B. Smith, M. Bresciani, T. Egerton, C. Giardino, C., L. Lin, T. Moore, A. Ruiz-Verdu, S. Ruberg, S.G.H. Simis, R. Stumpf, and D. Vaiciute. 2021. Advancing cyanobacteria biomass estimation from hyperspectral observations: Demonstrations with HICO and PRISMA imagery. Remote Sensing of Environment 266:112693. https://doi.org/10.1016/j.rse.2021.112693
Pahlevan, N., B. Smith, C. Binding, D. Gurlin, L. Li, M. Bresciani, and C. Giardino. 2021. Hyperspectral retrievals of phytoplankton absorption and chlorophyll-a in inland and nearshore coastal waters. Remote Sensing of Environment 253:112200. https://doi.org/10.1016/j.rse.2020.112200
Queally, N., Z. Ye, T. Zheng, A. Chlus, F. Schneider, R.P. Pavlick, and P.A. Townsend. 2022. FlexBRDF: A Flexible BRDF Correction for Grouped Processing of Airborne Imaging Spectroscopy Flightlines. Journal of Geophysical Research: Biogeosciences 127:e2021JG006622. https://doi.org/10.1029/2021JG006622
Soenen, S.A., D.R. Peddle, and C.A. Coburn. 2005. SCS+ C: A modified sun-canopy-sensor topographic correction in forested terrain. IEEE Transactions on geoscience and remote sensing 43:2148-2159. https://doi.org/10.1109/TGRS.2005.852480
Thompson, D.R., V. Natraj, R.O. Green, M.C. Helmlinger, B.C. Gao, and M.L. Eastwood. 2018. Optimal estimation for imaging spectrometer atmospheric correction. Remote Sensing of Environment 216, 355-373. https://doi.org/10.1016/j.rse.2018.07.003
Thompson D.R. 2021a. EMIT Manually Adjusted Vegetation Reflectance Spectra. Ecological Spectral Information System (EcoSIS). https://doi.org/10.21232/6sQDNjfv
Thompson D.R. 2021b. EMIT Manually Adjusted Surface Reflectance Spectra. Data set. Ecological Spectral Information System (EcoSIS). https://doi.org/10.21232/ezrQtdcw
Thompson D.R. 2021c. EMIT Manually Adjusted Snow and Liquids Reflectance Spectra. Data set. Ecological Spectral Information System (EcoSIS). https://doi.org/10.21232/xhgtM3A9
Townsend, P., M.M. Gierach, P.G. Brodrick, A.M. Chlus, H. Hua, O. Kwoun, M.J. Lucas, N. Malarout, D.F. Moroni, S. Neely, W. Olson-Duvall, J.K. Pon, S. Shah, and D. Yu. 2023. SISTER: Composite Release ID (CRID) Product Generation Files, 2023. ORNL DAAC, Oak Ridge, Tennessee, USA. https://doi.org/10.3334/ORNLDAAC/2231
Wang, Z., A. Chlus, R. Geygan, Z. Ye, T. Zheng, A. Singh, J.J. Couture, J. Cavenderâ Bares, E.L. Kruger, and P.A. Townsend. 2020. Foliar functional traits from imaging spectroscopy across biomes in eastern North America. New Phytologist, 228:494-511. https://doi.org/10.1111/nph.16711
Dataset Revisions
|
Release Date |
Product Version |
Description |
|
2024-06-11 |
V004 |
Final SISTER Workflow Version 004 delivery for archive and publication. |
|
unpublished |
V003 |
SISTER Workflow Version 003 is unpublished. The ORNL DAAC is maintaining consistent workflow versioning as generated by the SISTER Science Team. |
|
2023-05-31 |
V002 |
Published Composite Release ID (CRID) of workflow runs V002 and V001. This publication is superseded by V004. |
|
unpublished |
V001 |
Version 001 was generated by the SISTER Science Team but not intended for archive delivery. The ORNL DAAC is maintaining consistent workflow versioning as generated by the SISTER Science Team. |