Documentation Revision Date: 2023-06-04
Dataset Version: 1
Summary
There are 432 files with this dataset which includes three files in cloud optimized GeoTIFF format (.tif) for each of 24 flightlines. The geoTIFF files provide vegetative biochemical traits (nitrogen concentration, leaf mass area, and chlorophyll content). Additional files for each flightline include a quicklook image as well as processing product generation information for experimental reproducibility and a Product Generation Executable (PGE) log file.
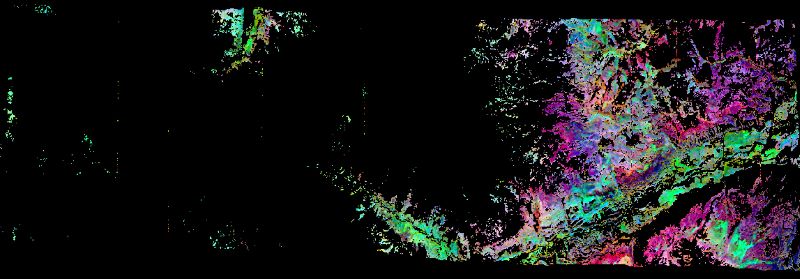
Figure 1. Portion of quicklook image for vegetation traits (chlorophyll content, nitrogen content, leaf mass per area) derived from AVIRIS-Classic imagery acquired on June 9 2011 north of Telluride, Colorado. North is to the right of the image. Source: SISTER_AVCL_L2B_VEGBIOCHEM_20110609T190737_001
Citation
Townsend, P., M.M. Gierach, C. Ade, A.M. Chlus, H. Hua, O. Kwoun, M.J. Lucas, N. Malarout, D.F. Moroni, S. Neely, W. Olson-Duvall, J.K. Pon, S. Shah, and D. Yu. 2023. SISTER: AVIRIS Classic L2B Vegetative Biochemical Traits 30 m V001. ORNL DAAC, Oak Ridge, Tennessee, USA. https://doi.org/10.3334/ORNLDAAC/2170
Table of Contents
- Dataset Overview
- Data Characteristics
- Application and Derivation
- Quality Assessment
- Data Acquisition, Materials, and Methods
- Data Access
- References
Dataset Overview
This dataset contains Level 2B vegetative biochemical traits (nitrogen concentration, leaf mass area, and chlorophyll content) at 30-m spatial resolution derived from data measured by the Airborne Visible/Infrared Imaging Spectrometer-Classic (AVIRIS or AVIRIS-CL) instrument during various campaigns (https://aviris.jpl.nasa.gov/). For the purposes of SISTER, only a handful of scenes have been selected from these campaigns, with a temporal range between 2011-05-26 and 2021-03-26 and spatial coverage entirely within the United States of America.
Project: SISTER
The Space-based Imaging Spectroscopy and Thermal pathfindER (SISTER) is a NASA project aimed at prototyping workflows and generating SBG-like data products in efforts to sustain and build the community to increase prospects for major scientific discovery post launch.
These data are associated with experimental products run by the SISTER Science Team as pre-launch modeling tools and data for algorithm development are investigated.The SISTER files for the Composite Release ID (CRID) 001 experimental run contain 19 separate collections (Table 1) that include four instruments and five data products (with the exception of the DESIS instrument). The output range for all sensors except DESIS is 400-2500 nm, while the DESIS output range is 400-990 nm.
Table 1. Summary of SISTER Sensors, Products, and Coding for all CRID 001 outputs available as separate datasets.
| Sensor | Product | Sensor_Level_Product |
| AVIRIS Classic | Resampled Surface Reflectance and Uncertainty | AVCL_L2A_RSRFL |
| Corrected Surface Reflectance | AVCL_L2A_CORFL | |
| Fractional Cover | AVCL_L2B_FRCOV | |
| Vegetative Biochemical Traits | AVCL_L2B_VEGBIOCHEM | |
| Snow Grain Size | AVCL_L2B_SNOWGRAIN | |
| AVIRIS Next Gen | Resampled Surface Reflectance and Uncertainty | AVNG_L2A_RSRFL |
| Corrected Surface Reflectance | AVNG_L2A_CORFL | |
| Fractional Cover | AVNG_L2B_FRCOV | |
| Vegetative Biochemical Traits | AVNG_L2B_VEGBIOCHEM | |
| Snow Grain Size | AVNG_L2B_SNOWGRAIN | |
| DESIS | Resampled Surface Reflectance and Uncertainty | DESIS_L2A_RSRFL |
| Corrected Surface Reflectance | DESIS_L2A_CORFL | |
| Fractional Cover | DESIS_L2B_FRCOV | |
| Vegetative Biochemical Traits | DESIS_L2B_VEGBIOCHEM | |
| PRISMA | Resampled Surface Reflectance and Uncertainty | PRISMA_L2A_RSRFL |
| Corrected Surface Reflectance | PRISMA_L2A_CORFL | |
| Fractional Cover | PRISMA_L2B_FRCOV | |
| Vegetative Biochemical Traits | PRISMA_L2B_VEGBIOCHEM | |
| Snow Grain Size | PRISMA_L2B_SNOWGRAIN |
Related Datasets:
See SISTER datasets with the Composite Release ID (CRID) version 001 (filter eg: V001)
Townsend, P., M.M. Gierach, P.G. Brodrick, A.M. Chlus, H. Hua, O. Kwoun, M.J. Lucas, N. Malarout, D.F. Moroni, S. Neely, W. Olson-Duvall, J.K. Pon, S. Shah, and D. Yu. 2023. SISTER: Composite Release ID (CRID) Product Generation Files, 2023. ORNL DAAC, Oak Ridge, Tennessee, USA. https://doi.org/10.3334/ORNLDAAC/2231
- SISTER_CRID_001.json - SISTER Composite Release ID (CRID) file contains details of repositories and versions used for this V001 run.
Data Characteristics
Spatial Coverage: Selected scenes/flight lines across the globe
Spatial Resolution: 30 m
Temporal Resolution: One-time estimates
Temporal Coverage: 2011-05-13 to 2021-03-26
Site Boundaries: Latitude and longitude are given in decimal degrees.
| Site | Westernmost Longitude | Easternmost Longitude | Northernmost Latitude | Southernmost Latitude |
|---|---|---|---|---|
| AVIRIS-Classic Flight Lines | -158.0554 | -107.6219 | 39.2655 | 20.9741 |
Data File Information
There are 432 files with this dataset which includes three files in cloud optimized GeoTIFF format (.tif) for each of 24 flightlines. The GeoTIFF files provide data for chlorophyll content, nitrogen concentration, leaf mass per area. Additional files for each flightline include a quicklook image as well as processing product generation information for experimental reproducibility and a Product Generation Executable (PGE) log file. In all, there are 18 files per flight line.
Data File Naming Conventions
File naming convention:
SISTER_<instrument>_<processing_level>_<product>_<flight_id>_<ver>_<var>.<ext>, where
where:
- <instrument> = the spectroscopy instrument that provided input radiance (Table 3)
- <processing_level> = the NASA Earthdata Data Processing Level
- <product> = the SISTER Project data product (Table 2)
- <flight_id> = flight line identifier, <YYMMDD>T<HHMMSS>, encoding the date and time by year (YY), month (MM), day (DD), and the UTC hour, minute, and second for the start of flight.
- <ver> = the SISTER processing version, also known as the CRID
- <var> = individual variable or measured parameter (CHL, LMA NIT)
- <ext> = file extension
<var>: vegetation biological traits variable description
- CHL (Chlorophyll):
- Band 1:chl_mean; chlorophyll content mean (micrograms per centimeter squared)
- Band 2: chl_std_dev; chlorophyll content standard deviation (micrograms per centimeter squared)
- Band 3: chl_qa_mask; quality assurance mask
- LMA (Leaf Mass per Area)
- Band 1: lma_mean; leaf mass per area mean (grams per meter squared)
- Band 2: lma_std_dev; leaf mass per area standard deviation (grams per meter squared)
- Band 3: lma_qa_mask; quality assurance mask
- NIT (Nitrogen Concentration)
- Band 1: nit_mean; nitrogen concentration mean (milligrams per gram)
- Band 2: nit_std_dev; leaf mass per area standard deviation (milligrams per gram)
- Band 3: nit_qa_mask; quality assurance mask
Example file name:
SISTER_AVCL_L2B_VEGBIOCHEM_20110513T175417_001_CHL.tif
Table 2. The outputs of the L2B vegetative biological traits PGE use the following naming convention and produce the following data products. The naming convention for these files follow the same pattern as described above.
|
Product description |
Example filename |
| Variable level 3-band geoTIFF file with variable, standard deviation, and quality assurance mask. |
SISTER_AVCL_L2B_VEGBIOCHEM_20210326T193449_001_<var>.tif |
| Variable level metadata including sensor, start and end time, description, bounding box, product, and processing level. |
SISTER_AVCL_L2B_VEGBIOCHEM_20210326T193449_001_<var>.met.json |
| Variable level dataset file version (e.g. {"version": "v1.0"}) used internally by SISTER to track product versions. Note that this is not the same as the CRID |
SISTER_AVCL_L2B_VEGBIOCHEM_20210326T193449_001_<var>.dataset.json |
| Variable level PGE context: supports data product traceability by providing provenance details, arguments, and runtime settings used during the run of the Product Generation Executable (PGE) on the SISTER platform |
SISTER_AVCL_L2B_VEGBIOCHEM_20210326T193449_001_<var>.context.json |
| Metadata including sensor, start and end time, description, bounding box, product, and processing level |
SISTER_AVCL_L2B_VEGBIOCHEM_20210326T193449_001.met.json |
| Dataset file version (e.g. {"version": "v1.0"}) used internally by SISTER to track product versions. Note that this is not the same as the CRID |
SISTER_AVCL_L2B_VEGBIOCHEM_20210326T193449_001.dataset.json |
| PGE context: supports data product traceability by providing provenance details, arguments, and runtime settings used during the run of the Product Generation Executable (PGE) on the SISTER platform. |
SISTER_AVCL_L2B_VEGBIOCHEM_20210326T193449_001.context.json |
| Quicklook image | SISTER_AVCL_L2B_VEGBIOCHEM_20210326T193449_001.png |
| PGE runconfig: defines inputs for the dataset's runs (one file per flight line) |
SISTER_AVCL_L2B_VEGBIOCHEM_20210326T193449_001.runconfig.json |
| PGE log (one file per flight line) |
SISTER_AVCL_L2B_VEGBIOCHEM_20210326T193449_001.log |
geoTIFF NoData values = -9999
Table 3. Possible SISTER Project Instruments available in the v001 dataset
| Instrument | Instrument fullname |
|---|---|
| AVCL | AVIRIS Classic |
| AVNG | AVIRIS Next Generation |
| DESIS | DLR Earth Sensing Imaging Spectrometer |
| PRISMA | PRecursore IperSpettrale della Missione Applicativa |
Application and Derivation
The 2018 National Academies’ Decadal Survey entitled, “Thriving on Our Changing Planet: A Decadal Strategy for Earth Observation from Space.” identified Surface Biology and Geology (SBG) as a Designated Observable (DO) with the following observing priorities:
* Terrestrial vegetation physiology, functional traits, and health
* Inland and coastal aquatic ecosystems physiology, functional traits, and health
* Snow and ice accumulation, melting, and albedo
* Active surface changes (eruptions, landslides, evolving landscapes, hazard risks)
* Effects of changing land use on surface energy, water, momentum, and C fluxes
* Managing agriculture, natural habitats, water use/quality, and urban development
To accomplish these priorities, the DO requires the combined use of visible to shortwave infrared (VSWIR) imaging spectroscopy and multispectral or hyperspectral thermal infrared (TIR) imagery acquired globally with sub-monthly temporal revisits over terrestrial, freshwater, and coastal marine habitats.
This approach presents some interesting challenges. Due to the high spatial and spectral resolution, SBG is expected to generate roughly 90 TB of data products per day. In addition, the number of existing community algorithms for processing this data is large and needs to be evaluated, and there is no particular consensus yet on standard file formats for hyperspectral data.
To help address a subset of these challenges the SBG Algorithms Working Group was formed to review and evaluate the algorithms applicable to the SBG DO. Also, the SISTER activity was created to prototype workflows and generate SBG-like data products.
This collection is one of several that include the first experimental data products produced by SISTER with the objective of generating SBG-like VSWIR imagery with 30-m spatial resolution and 10-nm spectral resolution. The scenes in the various SISTER collections include examples from the terrestrial, aquatic, snow/ice, and geologic domains, as well as from validation sites. The main purpose of these data are to provide the scientific community with prototype data products from the SBG workflow in order to get early feedback on things like useability, file formats, and metadata.
Quality Assessment
These products are still considered experimental, and no quality assessment procedure was conducted for this collection.
Data Acquisition, Materials, and Methods
The datasets in this collection were collected by the AVIRIS Classic (AVIRIS-CL) imaging spectrometer as part of various flight campaigns in the western United States over vegetation, aquatic and snow targets. AVIRIS-CL measures radiance at 10-nanometer (nm) intervals in the visible to shortwave infrared spectral range between 400 and 2500 nm. The raw AVIRIS data were processed to orthocorrected calibrated radiance by the Imaging Spectroscopy Group at JPL and then ingested into the SISTER platform for further downstream processing.
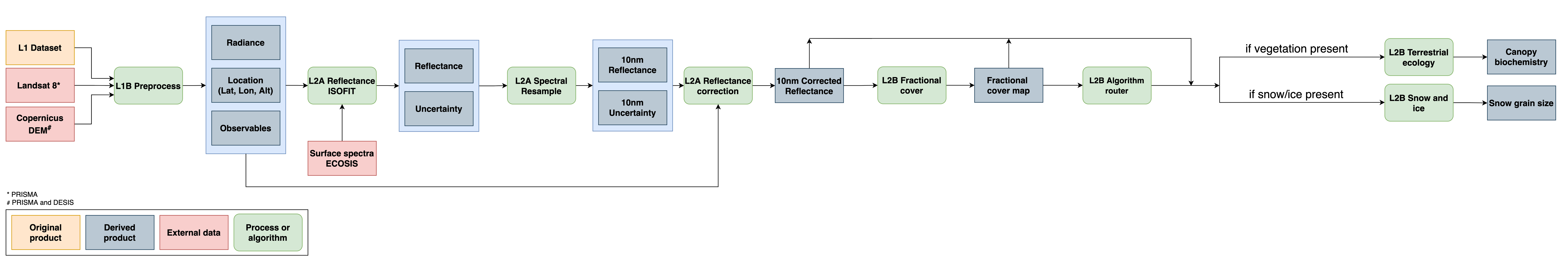
Figure 2. Workflow diagram of the SISTER 001 production run (Click on image to view full-resolution version).
First, images were spatially downsampled to 30-meter resolution and processed to surface reflectance using an optimal estimation atmospheric correction algorithm, ISOFIT (Thompson et al., 2018) with an open-source neural-network-based emulator for modelling radiative transfer (Brodrick et al., 2021). Next, spectral resampling to a 10-nm sampling interval was performed in a two-step calculation. Bands were first aggregated and averaged to the closest resolution to the target interval then a piecewise cubic interpolator was used to interpolate the spectra to the target wavelength spacing.
Following spectral resampling a combination of topographic, bidirectional reflectance distribution function (BRDF) and glint correction algorithms were applied to each image. Topographic correction was performed using the Sun-Canopy-Sensor+C algorithm (Soenen et al., 2005), BRDF correction was performed using the FlexBRDF algorithm (Queally et al., 2022) and glint correction was performed using the method of Gao and Li (2021).
Fractional cover maps were generated from the corrected reflectance datasets using a spectral mixture analysis. Spectral unmixing was performed using a generic four endmember dataset to derive fractional cover estimates of soil, vegetation, water and snow/ice. To minimize the impact of intraclass brightness variability on unmixing results a brightness normalization was applied.
Terrestrial vegetation biochemistry was estimated using partial least squares regression (PLSR) models. Permuted PLSR models for estimating chlorophyll content, nitrogen concentration, and leaf mass per area were developed using coincident NEON AOP canopy spectra, downsampled to 10nm, and field data collected by Wang et al. (2020). Biochemical trait estimates were only calculated for pixels with greater than 50% vegetation cover. In addition to mean biochemical trait estimates, per-pixel uncertainties were calculated along with a quality assurance mask which flags pixels with trait estimates outside of the range of data used to build the model.
For all datasets in the SISTER 001 processing version (also known as the CRID), workflow components, versions, and links to source code are summarized in Table 4 as provided in the SISTER_CRID_001.json dataset file.
Table 4. Key SISTER workflow components, versions, and links to source code used for V001 production (Townsend et al., 2023).
| software | version | url |
|---|---|---|
| maap-api-nasa | 2.0 | https://gitlab.com/geospec/maap-api-nasa/-/tags/2.0 |
| sister-preprocess | 2.0.0 | https://github.com/EnSpec/sister-preprocess/releases/tag/2.0.0 |
| sister-isofit | 2.0.0 | https://gitlab.com/geospec/sister-isofit/-/releases/2.0.0 |
| sister-resample | 2.0.1 | https://github.com/EnSpec/sister-resample/releases/tag/2.0.1 |
| sister-reflect_correct | 2.0.0 | https://github.com/EnSpec/sister-reflect_correct/releases/tag/2.0.0 |
| sister-fractional-cover | 1.0.0 | https://gitlab.com/geospec/sister-fractional-cover/-/releases/1.0.0 |
| sister-algorithm_router | 1.0.0 | https://github.com/EnSpec/sister-algorithm_router/releases/tag/1.0.0 |
| sister-trait_estimate | 1.0.0 | https://github.com/EnSpec/sister-trait_estimate/releases/tag/1.0.0 |
| sister-grainsize | 1.0.0 | https://github.com/EnSpec/sister-grainsize/releases/tag/1.0.0 |
Data Access
These data are available through the Oak Ridge National Laboratory (ORNL) Distributed Active Archive Center (DAAC).
SISTER: AVIRIS Classic L2B Vegetative Biochemical Traits 30 m V001
Contact for Data Center Access Information:
- E-mail: uso@daac.ornl.gov
- Telephone: +1 (865) 241-3952
References
Brodrick, P.G., D.R. Thompson, J.E. Fahlen, M.L. Eastwood, C.M. Sarture, S.R. Lundeen, W. Olson-Duvall, N. Carmon, and R.O. Green. Generalized radiative transfer emulation for imaging spectroscopy reflectance retrievals, Remote Sensing of Environment, Volume 261, 2021, 112476, ISSN 0034-4257. https://doi.org/10.1016/j.rse.2021.112476
Gao, B.C., and R.R. Li. 2021. Correction of Sunglint Effects in High Spatial Resolution Hyperspectral Imagery Using SWIR or NIR Bands and Taking Account of Spectral Variation of Refractive Index of Water. Advances in Environmental and Engineering Research, 2(3), 1-15. https://doi.org/10.21926/aeer.2103017
Queally, N., Z. Ye, T. Zheng, A. Chlus, F. Schneider, R.P. Pavlick, and P.A. Townsend. 2022. FlexBRDF: A Flexible BRDF Correction for Grouped Processing of Airborne Imaging Spectroscopy Flightlines. Journal of Geophysical Research: Biogeosciences, 127(1), e2021JG006622. https://doi.org/10.1029/2021JG006622
Soenen, S.A., D.R. Peddle, and C.A. Coburn. 2005. SCS+ C: A modified sun-canopy-sensor topographic correction in forested terrain.IEEE Transactions on geoscience and remote sensing, 43(9), 2148-2159. https://doi.org/10.1109/TGRS.2005.852480
Thompson, D.R., V. Natraj, R.O. Green, M.C. Helmlinger, B.C. Gao, and M.L. Eastwood. 2018. Optimal estimation for imaging spectrometer atmospheric correction. Remote Sensing of Environment 216, 355-373.https://doi.org/10.1016/j.rse.2018.07.003
Townsend, P., M.M. Gierach, P.G. Brodrick, A.M. Chlus, H. Hua, O. Kwoun, M.J. Lucas, N. Malarout, D.F. Moroni, S. Neely, W. Olson-Duvall, J.K. Pon, S. Shah, and D. Yu. 2023. SISTER: Composite Release ID (CRID) Product Generation Files, 2023. ORNL DAAC, Oak Ridge, Tennessee, USA. https://doi.org/10.3334/ORNLDAAC/2231
Wang, Z., A. Chlus, R. Geygan, Z. Ye, T. Zheng, A. Singh, J.J. Couture, J. Cavenderâ Bares, E.L. Kruger, and P.A. Townsend. 2020. Foliar functional traits from imaging spectroscopy across biomes in eastern North America. New Phytologist, 228(2), pp.494-511. https://doi.org/10.1111/nph.16711